bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0817_orf1
Length=191
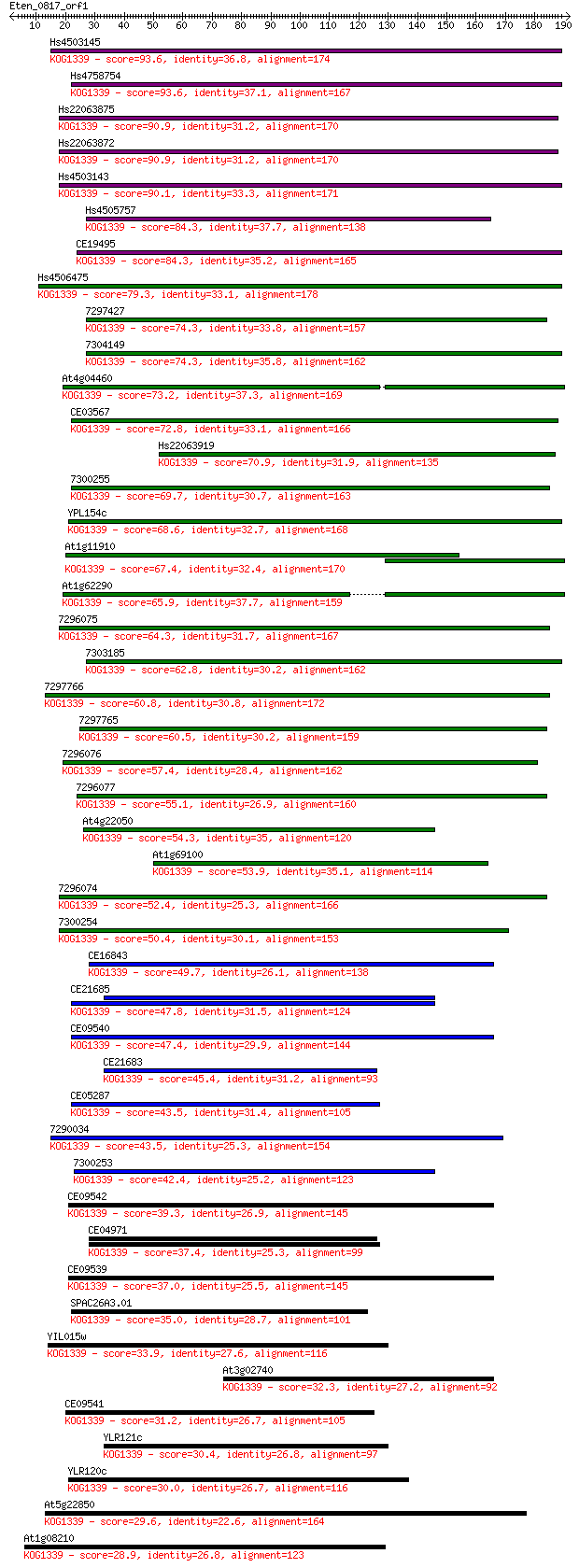
Score E
Sequences producing significant alignments: (Bits) Value
Hs4503145 93.6 2e-19
Hs4758754 93.6 2e-19
Hs22063875 90.9 1e-18
Hs22063872 90.9 1e-18
Hs4503143 90.1 2e-18
Hs4505757 84.3 1e-16
CE19495 84.3 1e-16
Hs4506475 79.3 4e-15
7297427 74.3 1e-13
7304149 74.3 1e-13
At4g04460 73.2 3e-13
CE03567 72.8 4e-13
Hs22063919 70.9 1e-12
7300255 69.7 4e-12
YPL154c 68.6 7e-12
At1g11910 67.4 2e-11
At1g62290 65.9 5e-11
7296075 64.3 1e-10
7303185 62.8 4e-10
7297766 60.8 2e-09
7297765 60.5 2e-09
7296076 57.4 2e-08
7296077 55.1 8e-08
At4g22050 54.3 2e-07
At1g69100 53.9 2e-07
7296074 52.4 5e-07
7300254 50.4 2e-06
CE16843 49.7 4e-06
CE21685 47.8 2e-05
CE09540 47.4 2e-05
CE21683 45.4 8e-05
CE05287 43.5 2e-04
7290034 43.5 3e-04
7300253 42.4 6e-04
CE09542 39.3 0.005
CE04971 37.4 0.020
CE09539 37.0 0.021
SPAC26A3.01 35.0 0.080
YIL015w 33.9 0.18
At3g02740 32.3 0.57
CE09541 31.2 1.3
YLR121c 30.4 2.2
YLR120c 30.0 2.6
At5g22850 29.6 3.8
At1g08210 28.9 6.3
> Hs4503145
Length=396
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 64/187 (34%), Positives = 94/187 (50%), Gaps = 24/187 (12%)
Query 15 GLPSSAL----PIVDQMVKEKVLDRNVFSVYMSEDIN--RPGEISFGAADPKYTFAGHTP 68
G PS A+ P+ D M+ + ++D +FSVYMS + E+ FG D + F+G +
Sbjct 188 GYPSLAVGGVTPVFDNMMAQNLVDLPMFSVYMSSNPEGGAGSELIFGGYDHSH-FSG-SL 245
Query 69 KWFPVISLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNV 128
W PV YW+I L +++ G E GC+A VDTG+SLITGPS I L A+
Sbjct 246 NWVPVTKQAYWQIALDNIQVGGTVMFCSE--GCQAIVDTGTSLITGPSDKIKQLQNAIGA 303
Query 129 AE-------NCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFM 181
A C+NL +P +TF + V ++L P Y + LD C++GF
Sbjct 304 APVDGEYAVECANLNVMPDVTFTING-----VPYTLSPTAYTL--LDFVDGMQFCSSGFQ 356
Query 182 AMDVPRP 188
+D+ P
Sbjct 357 GLDIHPP 363
> Hs4758754
Length=420
Score = 93.6 bits (231), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 62/177 (35%), Positives = 94/177 (53%), Gaps = 21/177 (11%)
Query 22 PIVDQMVKEKVLDRNVFSVYMSEDINRP--GEISFGAADPKYTFAGHTPKWFPVISLDYW 79
P +D +V++ +LD+ VFS Y++ D P GE+ G +DP + T + PV YW
Sbjct 201 PPMDVLVEQGLLDKPVFSFYLNRDPEEPDGGELVLGGSDPAHYIPPLT--FVPVTVPAYW 258
Query 80 EIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALN-----VAEN--- 131
+I + +K+ G +C K GC A +DTG+SLITGP+ I L A+ E
Sbjct 259 QIHMERVKV-GPGLTLCAK-GCAAILDTGTSLITGPTEEIRALHAAIGGIPLLAGEYIIL 316
Query 132 CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
CS + LP ++F+L ++ F+L DYV++ R C +GF A+DVP P
Sbjct 317 CSEIPKLPAVSFLLGGVW-----FNLTAHDYVIQT--TRNGVRLCLSGFQALDVPPP 366
> Hs22063875
Length=325
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 53/178 (29%), Positives = 91/178 (51%), Gaps = 23/178 (12%)
Query 18 SSALPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFPVISLD 77
S A P+ D + + ++ +++FSVY+S D + FG D Y + G W PV
Sbjct 130 SGATPVFDNIWNQGLVSQDLFSVYLSADDQSGSVVIFGGIDSSY-YTGSL-NWVPVTVEG 187
Query 78 YWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAEN------ 131
YW+I + + +NG++ E GC+A VDTG+SL+TGP+S I + + +EN
Sbjct 188 YWQITVDSITMNGEAIACAE--GCQAIVDTGTSLLTGPTSPIANIQSDIGASENSDGDMV 245
Query 132 --CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPR 187
CS + +LP + F + V + + P Y+++ + +C +GF M++P
Sbjct 246 VSCSAISSLPDIVFTING-----VQYPVPPSAYILQ------SEGSCISGFQGMNLPT 292
> Hs22063872
Length=325
Score = 90.9 bits (224), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 53/178 (29%), Positives = 91/178 (51%), Gaps = 23/178 (12%)
Query 18 SSALPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFPVISLD 77
S A P+ D + + ++ +++FSVY+S D + FG D Y + G W PV
Sbjct 130 SGATPVFDNIWNQGLVSQDLFSVYLSADDQSGSVVIFGGIDSSY-YTGSL-NWVPVTVEG 187
Query 78 YWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAEN------ 131
YW+I + + +NG++ E GC+A VDTG+SL+TGP+S I + + +EN
Sbjct 188 YWQITVDSITMNGEAIACAE--GCQAIVDTGTSLLTGPTSPIANIQSDIGASENSDGDMV 245
Query 132 --CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPR 187
CS + +LP + F + V + + P Y+++ + +C +GF M++P
Sbjct 246 VSCSAISSLPDIVFTING-----VQYPVPPSAYILQ------SEGSCISGFQGMNLPT 292
> Hs4503143
Length=412
Score = 90.1 bits (222), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/181 (31%), Positives = 94/181 (51%), Gaps = 21/181 (11%)
Query 18 SSALPIVDQMVKEKVLDRNVFSVYMSEDINRP--GEISFGAADPKYTFAGHTPKWFPVIS 75
++ LP+ D ++++K++D+N+FS Y+S D + GE+ G D KY + G + + V
Sbjct 209 NNVLPVFDNLMQQKLVDQNIFSFYLSRDPDAQPGGELMLGGTDSKY-YKG-SLSYLNVTR 266
Query 76 LDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE----- 130
YW++ L +++ +C K GC A VDTG+SL+ GP + L KA+
Sbjct 267 KAYWQVHLDQVEV-ASGLTLC-KEGCEAIVDTGTSLMVGPVDEVRELQKAIGAVPLIQGE 324
Query 131 ---NCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPR 187
C + TLP +T L G+ + L P DY ++ ++ C +GFM MD+P
Sbjct 325 YMIPCEKVSTLPAITLKLG---GK--GYKLSPEDYTLKV--SQAGKTLCLSGFMGMDIPP 377
Query 188 P 188
P
Sbjct 378 P 378
> Hs4505757
Length=388
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 52/147 (35%), Positives = 78/147 (53%), Gaps = 17/147 (11%)
Query 27 MVKEKVLDRNVFSVYMS-EDINRPGEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLHG 85
MV+E L VFSVY+S + + G + FG D + G W PV YW+IG+
Sbjct 199 MVQEGALTSPVFSVYLSNQQGSSGGAVVFGGVDSS-LYTGQI-YWAPVTQELYWQIGIEE 256
Query 86 LKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE--------NCSNLGT 137
I G++ G C + GC+A VDTG+SL+T P ++ L++A E NC+++
Sbjct 257 FLIGGQASGWCSE-GCQAIVDTGTSLLTVPQQYMSALLQATGAQEDEYGQFLVNCNSIQN 315
Query 138 LPTLTFVLKDIYGRLVNFSLEPRDYVV 164
LP+LTF++ V F L P Y++
Sbjct 316 LPSLTFIING-----VEFPLPPSSYIL 337
> CE19495
Length=398
Score = 84.3 bits (207), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 58/178 (32%), Positives = 92/178 (51%), Gaps = 25/178 (14%)
Query 24 VDQMVKEKVLDRN-VFSVYMSEDIN---RPGEISFGAADPKYTFAGHTPKWFPVISLDYW 79
+DQ+ + +N +F+ ++S D N GEI+ DP + + G+ W P++S DYW
Sbjct 198 MDQIFANSAICKNQLFAFWLSRDANDITNGGEITLCETDPNH-YVGNI-AWEPLVSEDYW 255
Query 80 EIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVIN---------PLIKALNVAE 130
I L + I+G ++ + VDTG+SL+TGP+ VI PL E
Sbjct 256 RIKLASVVIDGTTY---TSGPIDSIVDTGTSLLTGPTDVIKKIQHKIGGIPLFNGEYEVE 312
Query 131 NCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
CS + +LP +TF L NF L+ +DY+++ + G + C +GFM MD+P P
Sbjct 313 -CSKIPSLPNITFNLGG-----QNFDLQGKDYILQMSNGNG-GSTCLSGFMGMDIPAP 363
> Hs4506475
Length=406
Score = 79.3 bits (194), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 59/189 (31%), Positives = 94/189 (49%), Gaps = 22/189 (11%)
Query 11 GQVEGLPSSALPIVDQMVKEKVLDRNVFSVYMSEDINRP----GEISFGAADPKYTFAGH 66
G +E PI D ++ + VL +VFS Y + D G+I G +DP++ + G+
Sbjct 197 GFIEQAIGRVTPIFDNIISQGVLKEDVFSFYYNRDSENSQSLGGQIVLGGSDPQH-YEGN 255
Query 67 TPKWFPVISLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL 126
+ +I W+I + G+ + G S +CE GC A VDTG+S I+G +S I L++AL
Sbjct 256 F-HYINLIKTGVWQIQMKGVSV-GSSTLLCED-GCLALVDTGASYISGSTSSIEKLMEAL 312
Query 127 NVAE-------NCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAG 179
+ C+ TLP ++F L G+ ++L DYV +E + + C
Sbjct 313 GAKKRLFDYVVKCNEGPTLPDISFHLG---GK--EYTLTSADYVFQE--SYSSKKLCTLA 365
Query 180 FMAMDVPRP 188
AMD+P P
Sbjct 366 IHAMDIPPP 374
> 7297427
Length=372
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 53/163 (32%), Positives = 84/163 (51%), Gaps = 23/163 (14%)
Query 27 MVKEKVLDRNVFSVYMSEDINRP--GEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLH 84
MV + ++D +VFS Y++ D GE+ FG +D T + P+ YW+ +
Sbjct 195 MVSQGLVDNSVFSFYLARDGTSTMGGELIFGGSDASLYSGALT--YVPISEQGYWQFTMA 252
Query 85 GLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE----NCSNLGTLPT 140
G I+G S +C+ C+A DTG+SLI P + L + LNV E +CS++ +LP
Sbjct 253 GSSIDGYS--LCDD--CQAIADTGTSLIVAPYNAYITLSEILNVGEDGYLDCSSVSSLPD 308
Query 141 LTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAM 183
+TF +I G NF L+P Y+++ + NC + F M
Sbjct 309 VTF---NIGG--TNFVLKPSAYIIQ------SDGNCMSAFEYM 340
> 7304149
Length=392
Score = 74.3 bits (181), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 58/175 (33%), Positives = 87/175 (49%), Gaps = 28/175 (16%)
Query 27 MVKEKVLDRNVFSVYMSEDINRP--GEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLH 84
M ++ ++ VFS Y++ D P GEI FG +DP + T + PV YW+I
Sbjct 201 MYEQGLISAPVFSFYLNRDPASPEGGEIIFGGSDPNHYTGEFT--YLPVTRKAYWQI--- 255
Query 85 GLKINGKSFGVCE--KRGCRAAVDTGSSLITGP---SSVIN------PLIKALNVAENCS 133
K++ S G + K GC+ DTG+SLI P ++ IN P+I V +C
Sbjct 256 --KMDAASIGDLQLCKGGCQVIADTGTSLIAAPLEEATSINQKIGGTPIIGGQYVV-SCD 312
Query 134 NLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
+ LP + FVL G+ F LE +DY++ A+ C +GFM +D+P P
Sbjct 313 LIPQLPVIKFVLG---GK--TFELEGKDYILRV--AQMGKTICLSGFMGLDIPPP 360
> At4g04460
Length=508
Score = 73.2 bits (178), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 41/110 (37%), Positives = 64/110 (58%), Gaps = 5/110 (4%)
Query 19 SALPIVDQMVKEKVLDRNVFSVYMSEDINRP--GEISFGAADPKYTFAGHTPKWFPVISL 76
++ P+ MV++ ++ +FS +++ + P GEI FG DPK+ HT + PV
Sbjct 206 NSTPVWYNMVEKGLVKEPIFSFWLNRNPKDPEGGEIVFGGVDPKHFKGEHT--FVPVTHK 263
Query 77 DYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL 126
YW+ + L+I GK G C K GC A D+G+SL+TGPS+VI + A+
Sbjct 264 GYWQFDMGDLQIAGKPTGYCAK-GCSAIADSGTSLLTGPSTVITMINHAI 312
Score = 39.7 bits (91), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 7/61 (11%)
Query 129 AENCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
A +C + ++P +TF I GR +F L P+DY+ + G + C +GF AMD+ P
Sbjct 424 AVDCGRVSSMPIVTF---SIGGR--SFDLTPQDYIFKI--GEGVESQCTSGFTAMDIAPP 476
Query 189 R 189
R
Sbjct 477 R 477
> CE03567
Length=444
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 55/177 (31%), Positives = 88/177 (49%), Gaps = 24/177 (13%)
Query 22 PIVDQMVKEKVLDRNVFSVYMSE--DINRPGEISFGAADPKYTFAGHTPKWFPVISLDYW 79
P+ + + ++K + N+FS +++ D GEI+FG D + T + PV YW
Sbjct 217 PVFNTLFEQKKVPSNLFSFWLNRNPDSEIGGEITFGGIDSRRYVEPIT--YVPVTRKGYW 274
Query 80 EIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVI---------NPLIKALNVAE 130
+ + K+ G C GC+A DTG+SLI GP + I PLIK +
Sbjct 275 QFKMD--KVVGSGVLGCSN-GCQAIADTGTSLIAGPKAQIEAIQNFIGAEPLIKGEYMI- 330
Query 131 NCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPR 187
+C + TLP ++FV+ FSL+ DYV++ ++G C +GFM +D+P
Sbjct 331 SCDKVPTLPPVSFVIGG-----QEFSLKGEDYVLKV--SQGGKTICLSGFMGIDLPE 380
> Hs22063919
Length=267
Score = 70.9 bits (172), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 43/143 (30%), Positives = 72/143 (50%), Gaps = 23/143 (16%)
Query 52 ISFGAADPKYTFAGHTPKWFPVISLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSL 111
+ FG D Y + G W PV YW+I + + +NG++ E GC+A VDTG+SL
Sbjct 108 VIFGGIDSSY-YTGSL-NWVPVTVEGYWQITVDSITMNGEAIACAE--GCQAIVDTGTSL 163
Query 112 ITGPSSVINPLIKALNVAEN--------CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYV 163
+TGP+S I + + +EN CS + +LP + F + V + + P Y+
Sbjct 164 LTGPTSPIANIQSDIGASENSDGDMVVSCSAISSLPDIVFTING-----VQYPVPPSAYI 218
Query 164 VEELDARGNANNCAAGFMAMDVP 186
++ + +C +GF M++P
Sbjct 219 LQ------SEGSCISGFQGMNLP 235
> 7300255
Length=395
Score = 69.7 bits (169), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 50/173 (28%), Positives = 88/173 (50%), Gaps = 22/173 (12%)
Query 22 PIVDQMVKEKVLDRNVFSVYMSEDIN--RPGEISFGAADPKYTFAGHTPKWFPVISLDYW 79
P+ + M ++++D VFS Y+ + + + GE+ FG D K F+G + + P+ YW
Sbjct 204 PLFESMCDQQLVDECVFSFYLKRNGSERKGGELLFGGVD-KTKFSG-SLTYVPLTHAGYW 261
Query 80 EIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSS---VINPLIKALNVAEN----- 131
+ L +++ G + + R +A DTG+SL+ P +IN L+ L + N
Sbjct 262 QFPLDVIEVAGTR--INQNR--QAIADTGTSLLAAPPREYLIINSLLGGLPTSNNEYLLN 317
Query 132 CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMD 184
CS + +LP + F++ F L+PRDYV+ + G ++ C + F MD
Sbjct 318 CSEIDSLPEIVFIIGG-----QRFGLQPRDYVMSATNDDG-SSICLSAFTLMD 364
> YPL154c
Length=405
Score = 68.6 bits (166), Expect = 7e-12, Method: Compositional matrix adjust.
Identities = 55/179 (30%), Positives = 79/179 (44%), Gaps = 27/179 (15%)
Query 21 LPIVDQMVKEKVLDRNVFSVYM---SEDINRPGEISFGAADPKYTFAGHTPKWFPVISLD 77
+P +++ +LD F+ Y+ S+D GE +FG D F G W PV
Sbjct 211 VPPFYNAIQQDLLDEKRFAFYLGDTSKDTENGGEATFGGIDES-KFKGDI-TWLPVRRKA 268
Query 78 YWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPS---SVINPLIKAL-----NVA 129
YWE+ G+ + G + E G AA+DTG+SLIT PS +IN I A
Sbjct 269 YWEVKFEGIGL-GDEYAELESHG--AAIDTGTSLITLPSGLAEMINAEIGAKKGWTGQYT 325
Query 130 ENCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
+C+ LP L F NF++ P DY +E + +C + MD P P
Sbjct 326 LDCNTRDNLPDLIFNFNG-----YNFTIGPYDYTLEV------SGSCISAITPMDFPEP 373
> At1g11910
Length=506
Score = 67.4 bits (163), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 41/136 (30%), Positives = 67/136 (49%), Gaps = 11/136 (8%)
Query 20 ALPIVDQMVKEKVLDRNVFSVYMSE--DINRPGEISFGAADPKYTFAGHTPKWFPVISLD 77
A P+ M+K+ ++ VFS +++ D GE+ FG DP + HT + PV
Sbjct 202 AAPVWYNMLKQGLIKEPVFSFWLNRNADEEEGGELVFGGVDPNHFKGKHT--YVPVTQKG 259
Query 78 YWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSNLGT 137
YW+ + + I G G CE GC A D+G+SL+ GP+++I + A+ A G
Sbjct 260 YWQFDMGDVLIGGAPTGFCES-GCSAIADSGTSLLAGPTTIITMINHAIGAA------GV 312
Query 138 LPTLTFVLKDIYGRLV 153
+ + D YG+ +
Sbjct 313 VSQQCKTVVDQYGQTI 328
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 7/61 (11%)
Query 129 AENCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
A +C+ L T+PT++ I G++ F L P +YV++ G C +GF+A+DV P
Sbjct 422 AVDCAQLSTMPTVSLT---IGGKV--FDLAPEEYVLKV--GEGPVAQCISGFIALDVAPP 474
Query 189 R 189
R
Sbjct 475 R 475
> At1g62290
Length=526
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 37/100 (37%), Positives = 56/100 (56%), Gaps = 5/100 (5%)
Query 19 SALPIVDQMVKEKVLDRNVFSVYMSED--INRPGEISFGAADPKYTFAGHTPKWFPVISL 76
+A P+ M+K+ ++ R VFS +++ D GEI FG DPK+ HT + PV
Sbjct 208 NATPVWYNMLKQGLIKRPVFSFWLNRDPKSEEGGEIVFGGVDPKHFRGEHT--FVPVTQR 265
Query 77 DYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPS 116
YW+ + + I G+S G C GC A D+G+SL+ GP+
Sbjct 266 GYWQFDMGEVLIAGESTGYC-GSGCSAIADSGTSLLAGPT 304
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 23/61 (37%), Positives = 35/61 (57%), Gaps = 7/61 (11%)
Query 129 AENCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
A +CS L +PT++F I G++ F L P +YV++ G C +GF A+D+P P
Sbjct 442 AVDCSQLSKMPTVSFT---IGGKV--FDLAPEEYVLK--IGEGPVAQCISGFTALDIPPP 494
Query 189 R 189
R
Sbjct 495 R 495
> 7296075
Length=410
Score = 64.3 bits (155), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 53/180 (29%), Positives = 86/180 (47%), Gaps = 30/180 (16%)
Query 18 SSALPIVDQMVKEKVLDRNVFSVYMSEDINRP---GEISFGAADPKYTFAGHTPKWFPVI 74
+S P ++ + +++R+VFS+Y++ + GE+ G +D ++G + PV
Sbjct 215 NSITPPFYNLMAQGLVNRSVFSIYLNRNGTNAINGGELILGGSDSG-LYSG-CLTYVPVS 272
Query 75 SLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSV---INPLIKALN-VAE 130
S YW+ + +NG F CE C A +D G+SLI P V IN ++ LN A
Sbjct 273 SAGYWQFTMTSANLNG--FQFCEN--CEAILDVGTSLIVVPEQVLDTINQILGVLNPTAS 328
Query 131 N------CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMD 184
N CS++G LP + F + F L+ DYV+ N C +GF +M+
Sbjct 329 NGVFLVDCSSIGDLPDIVFTVAR-----RKFPLKSSDYVLRY------GNTCVSGFTSMN 377
> 7303185
Length=404
Score = 62.8 bits (151), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 49/171 (28%), Positives = 84/171 (49%), Gaps = 21/171 (12%)
Query 27 MVKEKVLDRNVFSVYMSEDINRP-GEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLHG 85
M+++ +L + +FSVY+S + + G I FG ++P Y T + V YW++ +
Sbjct 213 MMEQGLLTKPIFSVYLSRNGEKDGGAIFFGGSNPHYYTGNFT--YVQVSHRAYWQVKMDS 270
Query 86 LKINGKSFGVCEKRGCRAAVDTGSSLITGP---SSVINPLIKAL-----NVAENCSNLGT 137
I ++ +C++ GC +DTG+S + P + +IN I C ++
Sbjct 271 AVI--RNLELCQQ-GCEVIIDTGTSFLALPYDQAILINESIGGTPSSFGQFLVPCDSVPD 327
Query 138 LPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAMDVPRP 188
LP +TF L GR F LE +YV D + C++ F+A+D+P P
Sbjct 328 LPKITFTLG---GR--RFFLESHEYVFR--DIYQDRRICSSAFIAVDLPSP 371
> 7297766
Length=391
Score = 60.8 bits (146), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 53/187 (28%), Positives = 83/187 (44%), Gaps = 28/187 (14%)
Query 13 VEGLPSSAL------PIVDQMVKEKVLDRNVFSVYMSEDIN--RPGEISFGAADPKYTFA 64
+ GL SA+ P D M+ + +LD V S Y+ R GE+ G D
Sbjct 184 ILGLAFSAIAVDGVTPPFDNMISQGLLDEPVISFYLKRQGTAVRGGELILGGIDSSLYRG 243
Query 65 GHTPKWFPVISLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIK 124
T + PV YW+ ++ +K NG +C GC+A DTG+SLI P + + +
Sbjct 244 SLT--YVPVSVPAYWQFKVNTIKTNGTL--LCN--GCQAIADTGTSLIAVPLAAYRKINR 297
Query 125 ALNVAEN-------CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCA 177
L +N C + +LP + +I G + F+L PRDY+V+ + C
Sbjct 298 QLGATDNDGEAFVRCGRVSSLPKVNL---NIGGTV--FTLAPRDYIVKV--TQNGQTYCM 350
Query 178 AGFMAMD 184
+ F M+
Sbjct 351 SAFTYME 357
> 7297765
Length=423
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 48/169 (28%), Positives = 80/169 (47%), Gaps = 22/169 (13%)
Query 25 DQMVKEKVLDRNVFSVYMSED--INRPGEISFGAADPKYTFAGHTPKWFPVISLDYWEIG 82
D ++++ ++ VFSVY+ D GE+ +G D + + G + PV YW+
Sbjct 199 DNIIRQGLVKHPVFSVYLRRDGTSQSGGEVIWGGID-RSIYRG-CINYVPVSMPAYWQFT 256
Query 83 LHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE--------NCSN 134
+ +KI G + GC+A DTG+SLI P + K LN + +CS+
Sbjct 257 ANSVKIEG----ILLCNGCQAIADTGTSLIAVPLRAYKAINKVLNATDAGDGEAFVDCSS 312
Query 135 LGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAM 183
L LP + +I G ++L P+DY+ ++ A N C +GF +
Sbjct 313 LCRLPNVNL---NIGG--TTYTLTPKDYIY-KVQADNNQTLCLSGFTYL 355
> 7296076
Length=405
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 46/171 (26%), Positives = 77/171 (45%), Gaps = 22/171 (12%)
Query 19 SALPIVDQMVKEKVLDRNVFSVYMSEDINRP--GEISFGAADPKYTFAGHTPKWFPVISL 76
+ +P + +E ++D VF Y++ + + G+++ G D AG + PV
Sbjct 213 NVVPPFYNLYEEGLIDEPVFGFYLARNGSAVDGGQLTLGGTDQN-LIAGEM-TYTPVTQQ 270
Query 77 DYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSV---INPLIKALNVAEN-- 131
YW+ ++ + NG GC+A DTG+SLI PS+ +N LI + + +
Sbjct 271 GYWQFAVNNITWNGTVI----SSGCQAIADTGTSLIAAPSAAYIQLNNLIGGVPIQGDYY 326
Query 132 --CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGF 180
CS + +LP LT + NF L P Y+ + GN C + F
Sbjct 327 VPCSTVSSLPVLTINIGG-----TNFYLPPSVYI--QTYTEGNYTTCMSTF 370
> 7296077
Length=418
Score = 55.1 bits (131), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 43/175 (24%), Positives = 81/175 (46%), Gaps = 34/175 (19%)
Query 24 VDQMVKEKVLDRNVFSVYM---SEDINRPGEISFGAADPKYTFAGHTPKWFPVISLDYWE 80
+D ++++ ++D +FS+Y+ + D + G + G +DP T + PV + +W+
Sbjct 217 LDNILEQGLIDEPIFSLYVNRNASDASNGGVLLLGGSDP--TLYSGCLTYVPVSKVGFWQ 274
Query 81 IGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE---------- 130
I + ++I K +C C+A D G+SLI P + + K L + E
Sbjct 275 ITVGQVEIGSKK--LCSN--CQAIFDMGTSLIIVPCPALKIINKKLGIKETDRKDGVYII 330
Query 131 NCSNLGTLPTLTFVL--KDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAM 183
+C + LP + F + KD F+L P DY++ + C +GF ++
Sbjct 331 DCKKVSHLPKIVFNIGWKD-------FTLNPSDYILNY------SGTCVSGFSSL 372
> At4g22050
Length=336
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 42/131 (32%), Positives = 65/131 (49%), Gaps = 22/131 (16%)
Query 26 QMVKEKVLDRNVFSVYM-----SEDINRPGEISFGAADPKYTFAGHT------PKWFPVI 74
MV + + +NVFS+++ S +IN GE+ FG P + HT P F +
Sbjct 171 SMVFQGKIAKNVFSIWLRRFSNSGEIN-GGEVVFGGIIPAHFSGDHTYVDVEGPGNFFAM 229
Query 75 SLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSN 134
S + W + GK+ +C GC+A VD+GSS I P + + + + V NC+N
Sbjct 230 S-NIW--------VGGKNTNICSS-GCKAIVDSGSSNINVPMDSADEIHRYIGVEPNCNN 279
Query 135 LGTLPTLTFVL 145
TLP +TF +
Sbjct 280 FETLPDVTFTI 290
> At1g69100
Length=343
Score = 53.9 bits (128), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 40/119 (33%), Positives = 57/119 (47%), Gaps = 12/119 (10%)
Query 50 GEISFGAADPKYTFAGHTPKWFPV-ISLDYWEIGLHGLKINGK-SFGVCEKRGCRAAVDT 107
G+I FG DPK H + P+ +S D W+I + + INGK + C+ C A VD+
Sbjct 178 GQIMFGGFDPKQFKGEHV--YVPMKLSDDRWKIKMSKIYINGKPAINFCDDVECTAMVDS 235
Query 108 GSSLITGPSSVINPLIK---ALNVAENCSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYV 163
GS+ I GP + + K A V C LP + F +I G+ + L DYV
Sbjct 236 GSTDIFGPDEAVGKIYKEIGATKVIIRCEQFPALPDIYF---EIGGK--HLRLTKHDYV 289
> 7296074
Length=393
Score = 52.4 bits (124), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 42/172 (24%), Positives = 80/172 (46%), Gaps = 17/172 (9%)
Query 18 SSALPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFPVISLD 77
+ ++P QM+ ++++++ +FS ++ + G + G ++ + T + V
Sbjct 200 TKSVPSFQQMIDQQLVEQPIFSFHLKSGSSDGGSMILGGSNSSLYYGPLT--YTNVTEAK 257
Query 78 YWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCS-NLG 136
YW L + ++GK + G +A +DTG+SLI GP + + K + N + NL
Sbjct 258 YWSFKLDFIAVHGKG-SRSSRTGNKAIMDTGTSLIVGPVLEVLYINKDIGAEHNKTYNLY 316
Query 137 T-----LPTLTFVLKDIYGRLVNFSLEPRDYVVEELDARGNANNCAAGFMAM 183
T +P L ++ I G+ F ++P YV+ N C + FM M
Sbjct 317 TVACESIPQLPIIVFGIAGK--EFFVKPHTYVIRY------DNFCFSAFMDM 360
> 7300254
Length=309
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 73/161 (45%), Gaps = 24/161 (14%)
Query 18 SSALPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFPVISLD 77
S+ P ++ + +++L++ VFSVY+ D P EI FG D F G + PV
Sbjct 122 SNTTPFLELLCAQRLLEKCVFSVYLRRD---PREIVFGGFDES-KFEGKL-HYVPVSQWH 176
Query 78 YWEIGLHGLKINGKSFGVCEKRG-CRAAVDTGSSLITGPSSVINPLIKALNVAEN----- 131
W L+I+ S G + G A +DTG+SL+ P + L+ L+
Sbjct 177 TWS-----LQISKSSVGTKQIGGKSNAILDTGTSLVLVPQQTYHNLLNTLSAKLQNGYFV 231
Query 132 --CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVEELDAR 170
C + G+LP + ++ D F L DY++E L R
Sbjct 232 VACKS-GSLPNINILIGDKV-----FPLTSSDYIMEVLLDR 266
> CE16843
Length=394
Score = 49.7 bits (117), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 36/151 (23%), Positives = 63/151 (41%), Gaps = 23/151 (15%)
Query 28 VKEKVLDRNVFSVYMSED-INRP---GEISFGAADPKYTFAGHTPKWFPVISLDYWEIGL 83
V++ ++D +F+VY+ I + G ++G DP + G W P+ YW+ +
Sbjct 203 VEQGLVDEPIFTVYLEHHGIKKEASGGYFTYGGEDPDH--CGEIITWIPLTKAAYWQFRM 260
Query 84 HGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAEN---------CSN 134
G+ I+ + K G DTG+S I GP VI + + CS
Sbjct 261 QGIGIDNVN---EHKNGWEVISDTGTSFIGGPGKVIQDIANKYGATYDEYSDSYVIPCSE 317
Query 135 LGTLPTLTFVLKDIYGRLVNFSLEPRDYVVE 165
+ L +L + D+ F ++P + V
Sbjct 318 IKNLRSLKMKINDL-----TFEIDPINLVAH 343
> CE21685
Length=829
Score = 47.8 bits (112), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 37/128 (28%), Positives = 58/128 (45%), Gaps = 22/128 (17%)
Query 33 LDRNVFSVYMSEDIN-----RPGEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLHGLK 87
LD VFSV++ I G I++G D K AG T + P+ + YW+ K
Sbjct 640 LDVPVFSVWLDRKIQASHGGSAGMITYGGIDTKNCDAGVT--YVPLTAKTYWQ-----FK 692
Query 88 INGKSFGVCEKRGCRAAV-DTGSSLITGPSSVINPLIKALNVAEN---------CSNLGT 137
++G + G + G + DTGSS I+ P ++IN + + + CS + T
Sbjct 693 MDGFAVGTYSQYGYNQVISDTGSSWISAPYAMINDIATQTHATWDEMNEIYTVKCSTMKT 752
Query 138 LPTLTFVL 145
P L F +
Sbjct 753 QPDLVFTI 760
Score = 41.6 bits (96), Expect = 9e-04, Method: Compositional matrix adjust.
Identities = 33/138 (23%), Positives = 61/138 (44%), Gaps = 20/138 (14%)
Query 22 PIVDQMVKEKVLDRNVFSVYMSEDIN-----RPGEISFGAADPKYTFAGHTPKWFPVISL 76
P + ++ +K LD +FS+++ + G I++GA D K A + + S
Sbjct 195 PPMQNLISQKQLDAPLFSIWVDRKLQVSQGGTGGLITYGAVDTKNCDA--QVNYVALSSK 252
Query 77 DYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAE------ 130
YW+ + G+ I + E +A DTGS+ + P+ V+N +++
Sbjct 253 TYWQFPMDGVAIGNYAMMKQE----QAISDTGSAWLGLPNPVLNAIVQQTKATYDWNYEI 308
Query 131 ---NCSNLGTLPTLTFVL 145
+CS + T P L F +
Sbjct 309 YTLDCSTMQTQPDLVFTI 326
> CE09540
Length=395
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 43/151 (28%), Positives = 73/151 (48%), Gaps = 15/151 (9%)
Query 22 PIVDQMVKEKVLDRNVFSVYMSEDINRP----GEISFGAADPKYTFAGHTPKWFPVISLD 77
P+++ + +K+LD+ +F+V++ G ++GA D T G + P+ S
Sbjct 198 PLIN-AINKKILDQPLFTVFLEHKGTAKNVGGGVFTYGAVDT--TNCGPVIAYQPLSSAT 254
Query 78 YWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSNLG- 136
Y++ G K+ S + + DTGSSLI GPS+VIN +AL + +
Sbjct 255 YYQFVADGFKLGSYS----NNKKSQVISDTGSSLIGGPSAVINGFAQALGAKYHSDDDAY 310
Query 137 TLPTLTF--VLKDIYGRLVNFSLEPRDYVVE 165
LP T L G V +S+EP +Y+++
Sbjct 311 YLPCSTSKGTLDITIGGNV-YSIEPVNYILD 340
> CE21683
Length=320
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 29/98 (29%), Positives = 45/98 (45%), Gaps = 11/98 (11%)
Query 33 LDRNVFSVYMSEDIN-----RPGEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLHGLK 87
LD VF+V+M + G I++GA D + P+ + YWE L G
Sbjct 205 LDAKVFTVWMDRKLQGSNGGNGGLITYGALDS--VNCADDISYVPLTAKTYWEFALDGFA 262
Query 88 INGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKA 125
+ S E + + DTG+S I P S+I+ ++KA
Sbjct 263 VGSFS----ETKTAKVISDTGTSWIGAPHSIISAVVKA 296
> CE05287
Length=428
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 11/110 (10%)
Query 22 PIVDQMVKEKVLDRNVFSVYM-----SEDINRPGEISFGAADPKYTFAGHTPKWFPVISL 76
P+ + M + K LD+ F VY+ + IN G + G D T W P+ +
Sbjct 235 PLFNLMNQGK-LDQPYFVVYLANIGPTSQING-GAFTVGGLDT--THCSSNVDWVPLSTQ 290
Query 77 DYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL 126
+W+ L G ++ S+ G +AA DT +S I P SV+ L KA+
Sbjct 291 TFWQFKLGG--VSSGSYSQAPNSGWQAAADTAASFIGAPKSVVTSLAKAV 338
> 7290034
Length=407
Score = 43.5 bits (101), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/163 (23%), Positives = 76/163 (46%), Gaps = 19/163 (11%)
Query 15 GLPSSAL----PIVDQMVKEKVLDRNVFSVYMSED---INRPGEISFGAADPKYTFAGHT 67
G PS A+ P M+++ ++ VFS ++ ++ GE+ G +DP ++G +
Sbjct 187 GFPSLAVDGVTPTFQNMMQQGLVQSPVFSFFLRDNGSVTFYGGELILGGSDPS-LYSG-S 244
Query 68 PKWFPVISLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALN 127
+ V+ YW+ +K+ S +A DTG+SLI P + + + + N
Sbjct 245 LTYVNVVQAAYWKFQTDYIKVGSTSISTF----AQAIADTGTSLIIAPQAQYDQISQLFN 300
Query 128 VAENCSNLGTLPTLTF--VLKDIYGRLVNFSLEPRDYVVEELD 168
N L + ++ ++ +I G V+F + + Y++EE D
Sbjct 301 A--NSEGLFECSSTSYPDLIINING--VDFKIPAKYYIIEEED 339
> 7300253
Length=465
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 31/131 (23%), Positives = 55/131 (41%), Gaps = 12/131 (9%)
Query 23 IVDQMVKEKVLDRNVFSVYMSEDINRPG--EISFGAADPKYTFAGHTPKWFPVISLDYWE 80
+V M E V+ F++ M + I FG+++ ++ + PV YW+
Sbjct 272 LVQNMCSEDVITSCKFAICMKGGGSSSRGGAIIFGSSNTSAYSGSNSYTYTPVTKKGYWQ 331
Query 81 IGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSN------ 134
L + + G +A VD+G+SLIT P+++ N + K + S
Sbjct 332 FTLQDIYVGGTKVS----GSVQAIVDSGTSLITAPTAIYNKINKVIGCRATSSGECWMKC 387
Query 135 LGTLPTLTFVL 145
+P TFV+
Sbjct 388 AKKIPDFTFVI 398
> CE09542
Length=389
Score = 39.3 bits (90), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 39/159 (24%), Positives = 69/159 (43%), Gaps = 28/159 (17%)
Query 21 LPIVDQMVKEKVLDRNVFSVYM----SEDINRPGEISFGAADPKYTFAGHTPKWFPVISL 76
+P + + + +LD+ +FSV++ + + G ++GA D T G + P+ S
Sbjct 194 VPPLINAINQGILDQPLFSVWLEHRGAANNVGGGVFTYGAIDT--TNCGALVAYQPLSSA 251
Query 77 DYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAENCSNL- 135
Y++ G K+ S + DTG+S + GP SV++ L KA + N
Sbjct 252 TYYQFKAAGFKLGSYS----NTKTVDVISDTGTSFLGGPQSVVDGLAKAAGATYDDFNEV 307
Query 136 ---------GTLPTLTFVLKDIYGRLVNFSLEPRDYVVE 165
GTL DI +S++P +Y+V+
Sbjct 308 YFIDCAAQPGTL--------DITIGTNTYSIQPVNYIVD 338
> CE04971
Length=709
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 47/98 (47%), Gaps = 7/98 (7%)
Query 28 VKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFPVISLDYWEIGLHGLK 87
V +K++D +F VY+ NR G+I+FG D + ++ P+ + ++ + +
Sbjct 188 VGQKLVDP-IFHVYLGNRPNRAGQITFGGQDTQN--CAKVKQYHPLTTGAAYQFYMTSVS 244
Query 88 INGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKA 125
S + G A DT SS+I GP +++ + A
Sbjct 245 SGAYS----ARNGWTALSDTASSIIYGPKAIVAGIADA 278
Score = 31.2 bits (69), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 22/103 (21%), Positives = 49/103 (47%), Gaps = 10/103 (9%)
Query 28 VKEKVLDRNVFSVYMSEDINRPGE----ISFGAADPKYTFAGHTPKWFPVISLDYWEIGL 83
+ + ++D +F+V++ + ++ G ++G D K G + P+ S Y++ +
Sbjct 518 INQNLVDLPLFTVFLEHEGDQNGVQGGVYTYGGIDTKN--CGPVIAYQPLSSATYYQFKM 575
Query 84 HGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL 126
+ S +G + DTG+SLI GP + ++ + A+
Sbjct 576 SAIG----SGSYHSSKGWQVISDTGTSLIGGPKAYVSAIADAV 614
> CE09539
Length=393
Score = 37.0 bits (84), Expect = 0.021, Method: Compositional matrix adjust.
Identities = 37/160 (23%), Positives = 72/160 (45%), Gaps = 30/160 (18%)
Query 21 LPIVDQMVKEKVLDRNVFSVYM-----SEDINRPGEISFGAADPKYTFAGHTPKWFPVIS 75
+P + + + +LD+ +F+V++ + D+ G ++GA D T G + P+ S
Sbjct 196 VPPLINAINQGLLDQPLFTVWLEHKGSANDVGG-GVFTYGAVDT--TNCGPVIAYQPLSS 252
Query 76 LDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAEN---- 131
Y++ K+ S + + DTG+S + GP +V+ L AL +
Sbjct 253 ATYYQFVADSFKLGSYS----NSKKYQVISDTGTSFLGGPKAVVAGLASALGATYHSNDD 308
Query 132 ------CSNLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVE 165
+N+GTL I G++ +S+ P +Y+V+
Sbjct 309 SYYVPCATNIGTLDIT------IGGKV--YSINPVNYIVD 340
> SPAC26A3.01
Length=514
Score = 35.0 bits (79), Expect = 0.080, Method: Compositional matrix adjust.
Identities = 29/114 (25%), Positives = 49/114 (42%), Gaps = 18/114 (15%)
Query 22 PIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFPVISLD---Y 78
IVDQ+V V+D F +Y++ED+ GE+ FG D K G + W + S D +
Sbjct 233 TIVDQLVSANVIDTPAFGIYLNEDV---GELIFGGYD-KAKING-SVHWVNISSSDDSTF 287
Query 79 WEIGLHGLKINGKSFG---VCEKRGCR-------AAVDTGSSLITGPSSVINPL 122
+ + L + + + KR + +DTG+ I P + +
Sbjct 288 YSVNLESITVTNSTSSNNVQSSKRSSKDIEVNTTVTLDTGTVYIYLPEDTVESI 341
> YIL015w
Length=587
Score = 33.9 bits (76), Expect = 0.18, Method: Composition-based stats.
Identities = 32/129 (24%), Positives = 60/129 (46%), Gaps = 16/129 (12%)
Query 14 EGLPSSALPIVDQMVK-EKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFAGHTPKWFP 72
EG P+ P Q++K EK++D +S++++ + G I FGA D + F+G FP
Sbjct 185 EGAPNEYYPNFPQILKSEKIIDVVAYSLFLNSPDSGTGSIVFGAID-ESKFSGDLFT-FP 242
Query 73 VISLDYWEI--GLHGLKINGKSFGVCEKRGCR----------AAVDTGSSLITGPSSVIN 120
+++ +Y I L + + G K C +D+G+SL+ P + +
Sbjct 243 MVN-EYPTIVDAPATLAMTIQGLGAQNKSSCEHETFTTTKYPVLLDSGTSLLNAPKVIAD 301
Query 121 PLIKALNVA 129
+ +N +
Sbjct 302 KMASFVNAS 310
> At3g02740
Length=488
Score = 32.3 bits (72), Expect = 0.57, Method: Compositional matrix adjust.
Identities = 25/108 (23%), Positives = 54/108 (50%), Gaps = 22/108 (20%)
Query 74 ISLDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKAL------- 126
++L+ E+G L+++ +F + +G +D+G++L+ P +V NPL+ +
Sbjct 286 VNLNAIEVGNSVLELSSNAFDSGDDKG--VIIDSGTTLVYLPDAVYNPLLNEILASHPEL 343
Query 127 ---NVAENCS------NLGTLPTLTFVLKDIYGRLVNFSLEPRDYVVE 165
V E+ + L PT+TF + + V+ ++ PR+Y+ +
Sbjct 344 TLHTVQESFTCFHYTDKLDRFPTVTFQ----FDKSVSLAVYPREYLFQ 387
> CE09541
Length=350
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 28/109 (25%), Positives = 50/109 (45%), Gaps = 10/109 (9%)
Query 20 ALPIVDQMVKEKVLDRNVFSVYMSEDINRP----GEISFGAADPKYTFAGHTPKWFPVIS 75
+P + +K+ +LD+ +FSV++ G ++GA D T + P+ S
Sbjct 151 VVPPLINAIKQGILDQPLFSVFLEHRGTLTNVGGGVFTYGAVDT--TNCEPVIGYQPLSS 208
Query 76 LDYWEIGLHGLKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIK 124
Y++ + ++ GK + DTG+S + GPS VIN + K
Sbjct 209 ATYFQFKANRFRL-GKYL---NAKVVDVISDTGTSFLGGPSVVINEIAK 253
> YLR121c
Length=508
Score = 30.4 bits (67), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 26/108 (24%), Positives = 50/108 (46%), Gaps = 14/108 (12%)
Query 33 LDRNVFSVYMSEDINRPGEISFGAAD-PKYTFAGHTPKWFPVISLDY---------WEIG 82
+D +S++++++ G I FGA D KY +T P+++L +++
Sbjct 207 IDATAYSLFLNDESQSSGSILFGAVDHSKYEGQLYT---IPLVNLYKSQGYQHPVAFDVT 263
Query 83 LHGLKINGKSFGVC-EKRGCRAAVDTGSSLITGPSSVINPLIKALNVA 129
L GL + + A +D+G++L PS + L K+LN +
Sbjct 264 LQGLGLQTDKRNITLTTTKLPALLDSGTTLTYLPSQAVALLAKSLNAS 311
> YLR120c
Length=569
Score = 30.0 bits (66), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 31/125 (24%), Positives = 57/125 (45%), Gaps = 12/125 (9%)
Query 21 LPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAAD-PKYTFAGHTPKWFPVISLD-- 77
PIV + + N +S+Y+++ G I FGA D KYT +T +S
Sbjct 279 FPIV--LKNSGAIKSNTYSLYLNDSDAMHGTILFGAVDHSKYTGTLYTIPIVNTLSASGF 336
Query 78 ----YWEIGLHGLKI--NGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALNVAEN 131
+++ ++G+ I +G S A +D+G++L P +V++ + L A+
Sbjct 337 SSPIQFDVTINGIGISDSGSSNKTLTTTKIPALLDSGTTLTYLPQTVVSMIATELG-AQY 395
Query 132 CSNLG 136
S +G
Sbjct 396 SSRIG 400
> At5g22850
Length=539
Score = 29.6 bits (65), Expect = 3.8, Method: Compositional matrix adjust.
Identities = 37/180 (20%), Positives = 77/180 (42%), Gaps = 20/180 (11%)
Query 13 VEGLPSSALPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGA-ADPKYTFAGHTPKWF 71
+ G + ++ Q+ + + R VFS + + G + G +P F P
Sbjct 225 IFGFGQQGMSVISQLASQGIAPR-VFSHCLKGENGGGGILVLGEIVEPNMVFTPLVPSQ- 282
Query 72 PVISLDYWEIGLHG--LKINGKSFGVCEKRGCRAAVDTGSSLITGPSSVINPLIKALN-- 127
P +++ I ++G L IN F +G +DTG++L + P ++A+
Sbjct 283 PHYNVNLLSISVNGQALPINPSVFSTSNGQG--TIIDTGTTLAYLSEAAYVPFVEAITNA 340
Query 128 VAENCSNLGTLPTLTFVLKDIYGRL-----VNFS------LEPRDYVVEELDARGNANNC 176
V+++ + + +V+ G + +NF+ L P+DY++++ + G A C
Sbjct 341 VSQSVRPVVSKGNQCYVITTSVGDIFPPVSLNFAGGASMFLNPQDYLIQQNNVGGTAVWC 400
> At1g08210
Length=566
Score = 28.9 bits (63), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 33/131 (25%), Positives = 59/131 (45%), Gaps = 16/131 (12%)
Query 6 RPGRSGQ-VEGLPSSALPIVDQMVKEKVLDRNVFSVYMSEDINRPGEISFGAADPKYTFA 64
RP R+ + GL +L ++ Q+ + + R VFS + D + G + G T
Sbjct 245 RPRRAVDGIFGLGQGSLSVISQLAVQGLAPR-VFSHCLKGDKSGGGIMVLGQIKRPDTV- 302
Query 65 GHTPKWFPVISLDYWEIGLHGLKINGK-------SFGVCEKRGCRAAVDTGSSLITGPSS 117
+TP V S ++ + L + +NG+ F + G +DTG++L P
Sbjct 303 -YTPL---VPSQPHYNVNLQSIAVNGQILPIDPSVFTIATGDG--TIIDTGTTLAYLPDE 356
Query 118 VINPLIKALNV 128
+P I+A++V
Sbjct 357 AYSPFIQAVSV 367
Lambda K H
0.319 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3202455494
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40