bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0811_orf1
Length=143
Score E
Sequences producing significant alignments: (Bits) Value
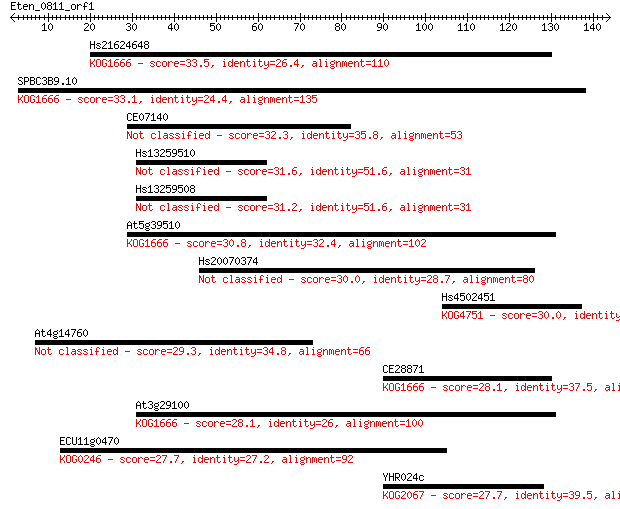
Hs21624648 33.5 0.12
SPBC3B9.10 33.1 0.16
CE07140 32.3 0.32
Hs13259510 31.6 0.55
Hs13259508 31.2 0.61
At5g39510 30.8 0.77
Hs20070374 30.0 1.5
Hs4502451 30.0 1.6
At4g14760 29.3 2.6
CE28871 28.1 5.1
At3g29100 28.1 6.4
ECU11g0470 27.7 7.1
YHR024c 27.7 8.5
> Hs21624648
Length=203
Score = 33.5 bits (75), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 29/122 (23%), Positives = 63/122 (51%), Gaps = 13/122 (10%)
Query 20 DTSRMPVLAQSKLSEIKDCLGAFEMELRSMSLAQRNCHLSQLQTCRNEFASLE---RRCL 76
+ +M + +L E K+ L ++E+R + R + +++++ + E LE +R
Sbjct 33 EKKQMVANVEKQLEEAKELLEQMDLEVREIPPQSRGMYSNRMRSYKQEMGKLETDFKRSR 92
Query 77 LAASGASKAP------TTTEGRQC---DHTLERLKNANVQLASTRKLAEETEEVGANILC 127
+A S + ++E ++ D+T ERL+ ++ +L + ++A ETE++G +L
Sbjct 93 IAYSDEVRNELLGDDGNSSENQRAHLLDNT-ERLERSSRRLEAGYQIAVETEQIGQEMLE 151
Query 128 NL 129
NL
Sbjct 152 NL 153
> SPBC3B9.10
Length=214
Score = 33.1 bits (74), Expect = 0.16, Method: Compositional matrix adjust.
Identities = 33/153 (21%), Positives = 71/153 (46%), Gaps = 20/153 (13%)
Query 3 VVAAEVESMLDD-HESGRDTSRMPVLAQSKLSEIKDCLGAFEMELRSMSLAQRNCHLSQL 61
++ A++E L+D +SG ++ + Q L+EI + +G E+E+ + ++R ++
Sbjct 10 LLRADIEEKLNDLSKSGENS--VIQSCQRLLNEIDEVIGQMEIEITGIPTSERGLVNGRI 67
Query 62 QTCRN-----------EFASLERRCLLAASGASKAPTTTEGRQCDH------TLERLKNA 104
++ R+ E +R+ L + + D RL+ +
Sbjct 68 RSYRSTLEEWRRHLKEEIGKSDRKALFGNRDETSGDYIASDQDYDQRTRLLQGTNRLEQS 127
Query 105 NVQLASTRKLAEETEEVGANILCNLYLPGRPLE 137
+ +L ++++A ETE +GA+IL +L+ LE
Sbjct 128 SQRLLESQRIANETEGIGASILRDLHGQRNQLE 160
> CE07140
Length=357
Score = 32.3 bits (72), Expect = 0.32, Method: Compositional matrix adjust.
Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 2/54 (3%)
Query 29 QSKLSEIKDCLGAFEMELRSMSLAQRNCHLSQLQTCRNEFASLERR-CLLAASG 81
++K+ ++ D L E++LR L+ N H S + C EF LE + CL SG
Sbjct 83 ETKMQKLVDALSYLEIKLRKFRLSAYNPHWSTI-PCLEEFLLLENKICLADKSG 135
> Hs13259510
Length=1278
Score = 31.6 bits (70), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 31 KLSEIKDCLGAFEMELRSMSLAQRNCHLSQL 61
K +E K A EMELR M +AQ N H+S L
Sbjct 558 KFAETKAHAKAIEMELRQMEVAQANRHMSLL 588
> Hs13259508
Length=1144
Score = 31.2 bits (69), Expect = 0.61, Method: Compositional matrix adjust.
Identities = 16/31 (51%), Positives = 19/31 (61%), Gaps = 0/31 (0%)
Query 31 KLSEIKDCLGAFEMELRSMSLAQRNCHLSQL 61
K +E K A EMELR M +AQ N H+S L
Sbjct 424 KFAETKAHAKAIEMELRQMEVAQANRHMSLL 454
> At5g39510
Length=221
Score = 30.8 bits (68), Expect = 0.77, Method: Compositional matrix adjust.
Identities = 33/125 (26%), Positives = 53/125 (42%), Gaps = 23/125 (18%)
Query 29 QSKLSEIKDCLGAFEMELRSMSLAQRN----------CHLSQLQTCRNEFASLERRC--- 75
+ KLSEIK L E+ +R M L R L + ++ N F + +R
Sbjct 35 KQKLSEIKSGLENAEVLIRKMDLEARTLPPNLKSSLLVKLREFKSDLNNFKTEVKRITSG 94
Query 76 ---------LLAASGASKAPTTTEGR-QCDHTLERLKNANVQLASTRKLAEETEEVGANI 125
LL A A + + R + + ERL ++ +R+ ETEE+G +I
Sbjct 95 QLNAAARDELLEAGMADTKTASADQRARLMMSTERLGRTTDRVKDSRRTMMETEEIGVSI 154
Query 126 LCNLY 130
L +L+
Sbjct 155 LQDLH 159
> Hs20070374
Length=458
Score = 30.0 bits (66), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/80 (28%), Positives = 37/80 (46%), Gaps = 4/80 (5%)
Query 46 LRSMSLAQRNCHLSQLQTCRNEFASLERRCLLAASGASKAPTTTEGRQCDHTLERLKNAN 105
L M+ Q++ + ++ FA++ LL A P TE D+TLE
Sbjct 193 LHGMAPQQKHGQQYKTKSSYKAFAAIPTNTLLLEQKALDEPAKTESVSKDNTLE----PP 248
Query 106 VQLASTRKLAEETEEVGANI 125
V+L +L ++TEE+ A I
Sbjct 249 VELYFPAQLRQQTEELCATI 268
> Hs4502451
Length=3418
Score = 30.0 bits (66), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 104 ANVQLASTRKLAEETEEVGANILCNLYLPGRPL 136
AN+QLA+T+K + V IL +Y P PL
Sbjct 3023 ANIQLAATKKTQYQQLPVSDEILFQIYQPREPL 3055
> At4g14760
Length=1676
Score = 29.3 bits (64), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 11/74 (14%)
Query 7 EVESMLDDHESGRDTSRMPVLAQSKLSEIKDCLGAFEMELRSMSLAQRNCHLS------- 59
EVE + D+E D+ RM + ++SE+ D G E E+R ++ N
Sbjct 1151 EVEELRKDYE---DSRRMRANLEWQISELSDVAGRQEEEIRKLNALNENLESEVQFLNKE 1207
Query 60 -QLQTCRNEFASLE 72
Q Q R E+ SLE
Sbjct 1208 IQRQQVREEYLSLE 1221
> CE28871
Length=224
Score = 28.1 bits (61), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 15/43 (34%), Positives = 26/43 (60%), Gaps = 3/43 (6%)
Query 90 EGRQCDHTL---ERLKNANVQLASTRKLAEETEEVGANILCNL 129
E RQ D + +RL+ + ++ ++A ETE++GA +L NL
Sbjct 121 EHRQEDQLIANTQRLERSTRKVQDAHRIAVETEQIGAEMLSNL 163
> At3g29100
Length=221
Score = 28.1 bits (61), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 26/100 (26%), Positives = 43/100 (43%), Gaps = 13/100 (13%)
Query 31 KLSEIKDCLGAFEMELRSMSLAQRNCHLSQLQTCRNEFASLERRCLLAASGASKAPTTTE 90
KL E K L F+ E++ ++ N T R+E L AS ++
Sbjct 73 KLREYKSDLNNFKTEVKRITSGNLNA------TARDELLEAGMADTLTASADQRSRLMMS 126
Query 91 GRQCDHTLERLKNANVQLASTRKLAEETEEVGANILCNLY 130
T +R+K++ R+ ETEE+G +IL +L+
Sbjct 127 TDHLGRTTDRIKDS-------RRTILETEELGVSILQDLH 159
> ECU11g0470
Length=581
Score = 27.7 bits (60), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 25/92 (27%), Positives = 36/92 (39%), Gaps = 11/92 (11%)
Query 13 DDHESGRDTSRMPVLAQSKLSEIKDCLGAFEMELRSMSLAQRNCHLSQLQTCRNEFASLE 72
D E G D L +K+C+ E + R + R L+Q+ +N F
Sbjct 399 DRKEMGSDVKNEGAEINKSLLALKECIRGMEKDKRHLPF--RQSKLTQI--LKNSFIGTS 454
Query 73 RRCLLAASGASKAPTTTEGRQCDHTLERLKNA 104
+ CL+A S P E HTL L+ A
Sbjct 455 KTCLIAT--ISPTPENVE-----HTLNTLRYA 479
> YHR024c
Length=482
Score = 27.7 bits (60), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 15/41 (36%), Positives = 20/41 (48%), Gaps = 3/41 (7%)
Query 90 EGRQ---CDHTLERLKNANVQLASTRKLAEETEEVGANILC 127
EGR C H L+RL + + R +AE E +G N C
Sbjct 53 EGRNLKGCTHILDRLAFKSTEHVEGRAMAETLELLGGNYQC 93
Lambda K H
0.315 0.127 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1639544670
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40