bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0801_orf1
Length=293
Score E
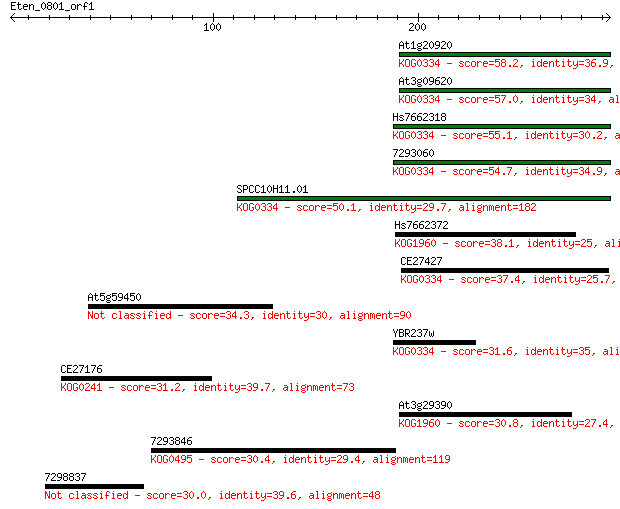
Sequences producing significant alignments: (Bits) Value
At1g20920 58.2 2e-08
At3g09620 57.0 4e-08
Hs7662318 55.1 2e-07
7293060 54.7 3e-07
SPCC10H11.01 50.1 6e-06
Hs7662372 38.1 0.022
CE27427 37.4 0.041
At5g59450 34.3 0.29
YBR237w 31.6 2.3
CE27176 31.2 2.7
At3g29390 30.8 3.6
7293846 30.4 4.9
7298837 30.0 5.8
> At1g20920
Length=1166
Score = 58.2 bits (139), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 59/103 (57%), Gaps = 6/103 (5%)
Query 191 ELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAPGVKFLHVEI 250
ELEIND+PQ AR+K+T KE + I E TGA + +GQ P + PG + L++ I
Sbjct 1070 ELEINDFPQNARWKVTHKETLGPISEWTGAAITTRGQFY-PTGR---IPGPGERKLYLFI 1125
Query 251 IAPTPIQVQRARHAVYELVESIALKSLNLSNTQRQAVGRYSVV 293
P+ V+ A+ + ++E I ++ +S+ A GRYSV+
Sbjct 1126 EGPSEKSVKHAKAELKRVLEDITNQA--MSSLPGGASGRYSVL 1166
> At3g09620
Length=989
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 35/103 (33%), Positives = 57/103 (55%), Gaps = 6/103 (5%)
Query 191 ELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAPGVKFLHVEI 250
ELEIND+PQ AR+K+T KE + I E +GA + +G+ E P + L++ +
Sbjct 893 ELEINDFPQNARWKVTHKETLGPISEWSGASITTRGKFYEAGRIP----GPEERKLYLFV 948
Query 251 IAPTPIQVQRARHAVYELVESIALKSLNLSNTQRQAVGRYSVV 293
PT I V+ A+ + ++E I ++ +L + GRYSV+
Sbjct 949 EGPTEISVKTAKAELKRVLEDITNQTFSLPGGAQS--GRYSVL 989
> Hs7662318
Length=1032
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 60/106 (56%), Gaps = 6/106 (5%)
Query 188 FVDELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAPGVKFLH 247
+ +ELEIND+PQ AR+K+T KE + RI E + A + ++G + P + + G + ++
Sbjct 933 YEEELEINDFPQTARWKVTSKEALQRISEYSEAAITIRGTYFPPGKEPK----EGERKIY 988
Query 248 VEIIAPTPIQVQRARHAVYELVESIALKSLNLSNTQRQAVGRYSVV 293
+ I + + VQ+A+ + L++ ++ N + Q GRY V+
Sbjct 989 LAIESANELAVQKAKAEITRLIKEELIRLQN--SYQPTNKGRYKVL 1032
> 7293060
Length=1224
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 37/106 (34%), Positives = 63/106 (59%), Gaps = 6/106 (5%)
Query 188 FVDELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAPGVKFLH 247
+ +ELEIND+PQ AR+K+T KE +++I E + A L V+G +V P K+ G + L+
Sbjct 1125 YEEELEINDFPQQARWKVTSKEALAQISEYSEAGLTVRGTYV-PQGKNPPD---GERKLY 1180
Query 248 VEIIAPTPIQVQRARHAVYELVESIALKSLNLSNTQRQAVGRYSVV 293
+ I + + + VQ+A+ + L++ LK + + + GRY VV
Sbjct 1181 LAIESCSELAVQKAKREITRLIKEELLKLSSAHHVFNK--GRYKVV 1224
> SPCC10H11.01
Length=1014
Score = 50.1 bits (118), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 54/185 (29%), Positives = 88/185 (47%), Gaps = 19/185 (10%)
Query 112 EKDPFMDAAAEALSILKQGTTASSSQLQTALDKIKAFSDSGELAAAISSVSAPSNPTLGK 171
E + ++ AEA S L++ T S+ T LD+++A + G A A ++ +A SN
Sbjct 846 EDEEDVETEAEAKSPLEKITPEKSTGDPT-LDRVRA-AVGGIAARAFANQTAQSNKLTQP 903
Query 172 VPGLSEKGFVCPETGNFVDELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEP 231
+ + G + ++EINDYPQ AR+ +T + + E TG + KG P
Sbjct 904 ISIIKTDG------DEYKAKMEINDYPQQARWAVTNNTNIVHVTELTGTSITTKGNFYLP 957
Query 232 ALKHRTQLAPGVKFLHVEIIAPTPIQVQRARHAVYELVESIALKSLNLS---NTQRQAVG 288
PG + L++ I P+ + V R A+ EL + L+ +N S + A G
Sbjct 958 G----KNPEPGEEKLYLWIEGPSELVVNR---AITEL-RRLLLEGINHSLEGGNKPSASG 1009
Query 289 RYSVV 293
RY+VV
Sbjct 1010 RYTVV 1014
> Hs7662372
Length=564
Score = 38.1 bits (87), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 46/88 (52%), Gaps = 2/88 (2%)
Query 189 VDELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAPGVKFLHV 248
V E+EIND P R +T+ + I +GA + +G+ + + + ++ PG + L++
Sbjct 104 VAEVEINDVPLTCRNLLTRGQTQDEISRLSGAAVSTRGRFM--TTEEKAKVGPGDRPLYL 161
Query 249 EIIAPTPIQVQRARHAVYELVESIALKS 276
+ T V RA + + E++ + +K+
Sbjct 162 HVQGQTRELVDRAVNRIKEIITNGVVKA 189
> CE27427
Length=970
Score = 37.4 bits (85), Expect = 0.041, Method: Compositional matrix adjust.
Identities = 26/101 (25%), Positives = 48/101 (47%), Gaps = 5/101 (4%)
Query 192 LEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAPGVKFLHVEII 251
+IND+PQ RYKI +E + + E + V+G HV P + + G + LH+ +
Sbjct 875 WDINDFPQQVRYKICSRESVGHVAELAEVGISVRGVHVPPGKEPKN----GERRLHLLLE 930
Query 252 APTPIQVQRARHAVYELVESIALKSLNLSNTQRQAVGRYSV 292
A + ++ A+ + +++ A + L + RY V
Sbjct 931 ARSERNLKAAKEEIIRIMKE-AFRQLTAQLQRGGTQARYKV 970
> At5g59450
Length=610
Score = 34.3 bits (77), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 27/90 (30%), Positives = 38/90 (42%), Gaps = 1/90 (1%)
Query 39 NSEKTKLTVDQQVEFMLQQALKGLPEADQAHYRDKLREAYKKHLSPRPKIQQKAAPGMQL 98
N KT T + +L Q + + DQ DKL+E + H S Q+ A
Sbjct 212 NKSKTHKTNTVDLRSLLTQCAQAVASFDQRRATDKLKEI-RAHSSSNGDGTQRLAFYFAE 270
Query 99 ALASSITGQAVPKEKDPFMDAAAEALSILK 128
AL + ITG P +PF + + ILK
Sbjct 271 ALEARITGNISPPVSNPFPSSTTSMVDILK 300
> YBR237w
Length=849
Score = 31.6 bits (70), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 0/40 (0%)
Query 188 FVDELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQ 227
F ++ IND PQ+ R++ T+ + I ETG + KG+
Sbjct 753 FYAKVYINDLPQIVRWEATKNTTLLFIKHETGCSITNKGK 792
> CE27176
Length=1595
Score = 31.2 bits (69), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 29/83 (34%), Positives = 40/83 (48%), Gaps = 11/83 (13%)
Query 26 PTPPSPPPGLPPV-------NSEKTK-LTVDQQVEFMLQQALKGLPEADQAHYRDKLREA 77
PTP + P GLPP S K+K Q+ E M ++LK L +AD H +REA
Sbjct 690 PTPITTPTGLPPFPFPANPKQSVKSKFFYWAQRKEEMFAESLKRL-KADVIHANALVREA 748
Query 78 --YKKHLSPRPKIQQKAAPGMQL 98
K L+ +PK Q +Q+
Sbjct 749 NMISKELNKKPKRQTTYDVTLQI 771
> At3g29390
Length=537
Score = 30.8 bits (68), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 41/86 (47%), Gaps = 9/86 (10%)
Query 191 ELEINDYPQVARYKITQKELMSRIMEETGAVLYVKGQHVEPALKHRTQLAP--GVKFLHV 248
E+ IND R+++T++ I TGAV+ +G K+R AP G K L++
Sbjct 87 EIVINDAEASLRHRLTKRSTQEDIQRSTGAVVITRG-------KYRPPNAPPDGEKPLYL 139
Query 249 EIIAPTPIQVQRARHAVYELVESIAL 274
I A +Q++ + + + A+
Sbjct 140 HISAAAQLQLKETTERILAVDRAAAM 165
> 7293846
Length=913
Score = 30.4 bits (67), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 35/123 (28%), Positives = 56/123 (45%), Gaps = 21/123 (17%)
Query 70 YRDK-LREAYKKHLSPRPKIQQKAAPGMQLALASSITGQAVPKEKDPFMDAAAEALSILK 128
YRD+ LRE +++ RPKIQQ+ + ++ +LA S+T + E +I +
Sbjct 123 YRDRRLREDLERYRQERPKIQQQFS-DLKRSLA-SVTSE--------------EWSTIPE 166
Query 129 QGTTASSSQLQTALDKIKAFSD---SGELAAAISSVSAPSNPTLGKVPGLSEKGFVCPET 185
G + + Q +K D S L SS PS+ VPG++ G + P T
Sbjct 167 VGDSRNRKQRNPRAEKFTPLPDSLISRNLGGESSSTLDPSSGLASMVPGVATPGMLTP-T 225
Query 186 GNF 188
G+
Sbjct 226 GDL 228
> 7298837
Length=1114
Score = 30.0 bits (66), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 19/48 (39%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query 18 AELAKRPPPTPPSPPPGLPPVNSEKTKLTVDQQVEFMLQQALKGLPEA 65
AELA R PT P P PV + L D ++ L KGLP+A
Sbjct 726 AELAARARPTEPPSTPNFKPVTTTDPSLFSDGTAKYYL--PTKGLPDA 771
Lambda K H
0.313 0.129 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 6531448310
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40