bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0775_orf3
Length=173
Score E
Sequences producing significant alignments: (Bits) Value
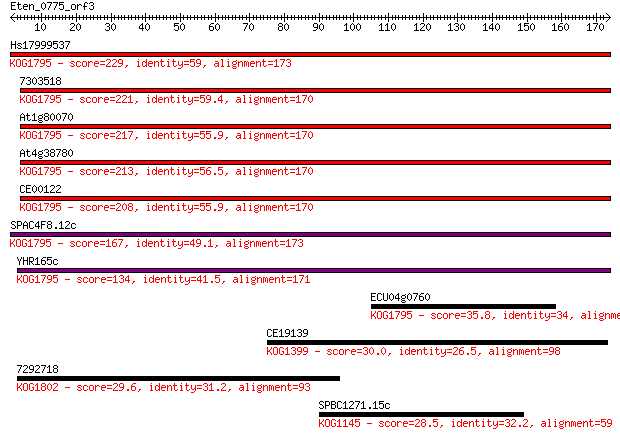
Hs17999537 229 2e-60
7303518 221 8e-58
At1g80070 217 8e-57
At4g38780 213 1e-55
CE00122 208 5e-54
SPAC4F8.12c 167 1e-41
YHR165c 134 7e-32
ECU04g0760 35.8 0.047
CE19139 30.0 2.3
7292718 29.6 3.4
SPBC1271.15c 28.5 7.0
> Hs17999537
Length=2335
Score = 229 bits (585), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 102/173 (58%), Positives = 126/173 (72%), Gaps = 2/173 (1%)
Query 1 QPPFLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCS 60
Q +L ++EPLGWIHTQPNE+PQL P+DV HAKI+ +N +WD ++TC+FTPGSC+
Sbjct 2165 QHEYLKEMEPLGWIHTQPNESPQLSPQDVTTHAKIMADNPSWDGEKTIIITCSFTPGSCT 2224
Query 61 LTAYRLTPQGYQWGKANRDMSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYN 120
LTAY+LTP GY+WG+ N D P GY H ER QMLLSD F GFF+VPA+ WNYN
Sbjct 2225 LTAYKLTPSGYEWGRQNTDKGNNPKGYLPSHYERVQMLLSDRFLGFFMVPAQSS--WNYN 2282
Query 121 FMGVKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQQEADVAEREDLFA 173
FMGV+ P MKY L L PK+FYHE HRP+HFL F L++ E A+REDL+A
Sbjct 2283 FMGVRHDPNMKYELQLANPKEFYHEVHRPSHFLNFALLQEGEVYSADREDLYA 2335
> 7303518
Length=2396
Score = 221 bits (562), Expect = 8e-58, Method: Compositional matrix adjust.
Identities = 101/171 (59%), Positives = 122/171 (71%), Gaps = 3/171 (1%)
Query 4 FLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCSLTA 63
+L D+EPLGWIHTQPNE PQL P+D+ HAKI+ EN NWD V+TC+FTPGSCSLTA
Sbjct 2228 YLKDMEPLGWIHTQPNELPQLSPQDITTHAKIMQENSNWDGEKTIVITCSFTPGSCSLTA 2287
Query 64 YRLTPQGYQWGKANRDMSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYNFMG 123
Y+LTP G++WG N D P GY H ER QMLLS+ F GFF+VPA+ WNYNFMG
Sbjct 2288 YKLTPSGFEWGSKNTDKGNNPKGYLPSHYERVQMLLSNKFLGFFMVPAQSS--WNYNFMG 2345
Query 124 VKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQ-QEADVAEREDLFA 173
V+ P MKY L L PK+FYHE HR +HFL F+ LE + A+RED++A
Sbjct 2346 VRHDPNMKYELQLANPKEFYHELHRTSHFLLFSNLEDGGDGAGADREDVYA 2396
> At1g80070
Length=2382
Score = 217 bits (553), Expect = 8e-57, Method: Compositional matrix adjust.
Identities = 95/170 (55%), Positives = 124/170 (72%), Gaps = 2/170 (1%)
Query 4 FLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCSLTA 63
FL DLEPLGW+HTQPNE PQL P+DV +H++IL NK WD ++TC+FTPGSCSLT+
Sbjct 2215 FLNDLEPLGWLHTQPNELPQLSPQDVTSHSRILENNKQWDGEKCIILTCSFTPGSCSLTS 2274
Query 64 YRLTPQGYQWGKANRDMSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYNFMG 123
Y+LT GY+WG+ N+D + P GY H E+ QMLLSD F GF++VP G WNY+F G
Sbjct 2275 YKLTQTGYEWGRLNKDNGSNPHGYLPTHYEKVQMLLSDRFLGFYMVPESGP--WNYSFTG 2332
Query 124 VKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQQEADVAEREDLFA 173
VK + +MKYS+ L +PK+FYHE HRP HFL+F+ +E+ + +RED F
Sbjct 2333 VKHTLSMKYSVKLGSPKEFYHEEHRPTHFLEFSNMEEADITEGDREDTFT 2382
> At4g38780
Length=2352
Score = 213 bits (542), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 96/170 (56%), Positives = 125/170 (73%), Gaps = 4/170 (2%)
Query 4 FLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCSLTA 63
FL DLEPLGWIHTQPNE PQL P+DV H ++L NK WDA ++TC+FTPGSCSLT+
Sbjct 2187 FLDDLEPLGWIHTQPNELPQLSPQDVTFHTRVLENNKQWDAEKCIILTCSFTPGSCSLTS 2246
Query 64 YRLTPQGYQWGKANRDMSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYNFMG 123
Y+LT GY+WG+ N+D + P GY H E+ QMLLSD F GF++VP G WNYNFMG
Sbjct 2247 YKLTQAGYEWGRLNKDTGSNPHGYLPTHYEKVQMLLSDRFFGFYMVPENGP--WNYNFMG 2304
Query 124 VKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQQEADVAEREDLFA 173
+ ++ YSL L TPK++YH+ HRP HFLQF+++E ++ D+ +R+D FA
Sbjct 2305 ANHTVSINYSLTLGTPKEYYHQVHRPTHFLQFSKME-EDGDL-DRDDSFA 2352
> CE00122
Length=2329
Score = 208 bits (529), Expect = 5e-54, Method: Compositional matrix adjust.
Identities = 95/171 (55%), Positives = 118/171 (69%), Gaps = 3/171 (1%)
Query 4 FLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCSLTA 63
L D EPLGW+HTQPNE PQL P+DV HAK+L +N +WD ++TC+FTPGS SLTA
Sbjct 2161 LLRDFEPLGWMHTQPNELPQLSPQDVTTHAKLLTDNISWDGEKTVMITCSFTPGSVSLTA 2220
Query 64 YRLTPQGYQWGKANRDMSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYNFMG 123
Y+LTP GY+WGKAN D P GY H E+ QMLLSD F G+F+VP+ G+WNYNF G
Sbjct 2221 YKLTPSGYEWGKANTDKGNNPKGYMPTHYEKVQMLLSDRFLGYFMVPS--NGVWNYNFQG 2278
Query 124 VKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQ-QEADVAEREDLFA 173
++SPAMK+ + L PK++YHE HRP HF F + A+RED FA
Sbjct 2279 QRWSPAMKFDVCLSNPKEYYHEDHRPVHFHNFKAFDDPLGTGSADREDAFA 2329
> SPAC4F8.12c
Length=2363
Score = 167 bits (422), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 85/174 (48%), Positives = 107/174 (61%), Gaps = 4/174 (2%)
Query 1 QPPFLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCS 60
QP L DLEPLGWIHTQ +E P L DV HAKIL+ + WD A +T ++ PGS S
Sbjct 2193 QPSILEDLEPLGWIHTQSSELPYLSSVDVTTHAKILSSHPEWDTK-AVTLTVSYIPGSIS 2251
Query 61 LTAYRLTPQGYQWGKANRDMSATPA-GYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNY 119
L AY ++ +G +WG N D+++ A GY AE+ Q+LLSD GFFLVP EG +WNY
Sbjct 2252 LAAYTVSKEGIEWGSKNMDINSDEAIGYEPSMAEKCQLLLSDRIQGFFLVPEEG--VWNY 2309
Query 120 NFMGVKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQQEADVAEREDLFA 173
NF G FSP M YSL LD P F+ HRP H + +T+LE + + D FA
Sbjct 2310 NFNGASFSPKMTYSLKLDVPLPFFALEHRPTHVISYTELETNDRLEEDMPDAFA 2363
> YHR165c
Length=2413
Score = 134 bits (338), Expect = 7e-32, Method: Composition-based stats.
Identities = 71/172 (41%), Positives = 101/172 (58%), Gaps = 7/172 (4%)
Query 3 PFLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCSLT 62
P LE LGWIHTQ E + +V H+K+ + K D ++ + TPGS SL+
Sbjct 2248 PDTEGLELLGWIHTQTEELKFMAASEVATHSKLFADKKR-DCIDISIFS---TPGSVSLS 2303
Query 63 AYRLTPQGYQWGKANRD-MSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYNF 121
AY LT +GYQWG+ N+D M+ G+ + AQ+LLSD +G F++P+ G +WNY F
Sbjct 2304 AYNLTDEGYQWGEENKDIMNVLSEGFEPTFSTHAQLLLSDRITGNFIIPS--GNVWNYTF 2361
Query 122 MGVKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQLEQQEADVAEREDLFA 173
MG F+ Y+ P +FY+E HRP HFLQF++L E AE+ D+F+
Sbjct 2362 MGTAFNQEGDYNFKYGIPLEFYNEMHRPVHFLQFSELAGDEELEAEQIDVFS 2413
> ECU04g0760
Length=2172
Score = 35.8 bits (81), Expect = 0.047, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 25/53 (47%), Gaps = 4/53 (7%)
Query 105 GFFLVPAEGGGIWNYNFMGVKFSPAMKYSLVLDTPKDFYHEAHRPAHFLQFTQ 157
G F VP +WNYNF + ++Y+ + P FY R HF +F Q
Sbjct 2113 GVFYVPE----MWNYNFARPFYDDRLEYTWKIGMPHGFYDGFCRVGHFSRFYQ 2161
> CE19139
Length=512
Score = 30.0 bits (66), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 26/103 (25%), Positives = 46/103 (44%), Gaps = 6/103 (5%)
Query 75 KANRDMSATPAGYSKGHAERAQMLLSDVFSGFFLVPAEGGGIWNYNFMGVKFSPAMKYSL 134
+ + + AG S + R +L +V F E GG+WNY + S MK S
Sbjct 3 ETKKQLLVVGAGASGLPSIRHALLYPNVEVTCFEKTNEVGGLWNYKPYKTELSTVMK-ST 61
Query 135 VLDTPKDFYHEAHRP-----AHFLQFTQLEQQEADVAEREDLF 172
V++T K+ + P A+F+ T++ + + A+ +L
Sbjct 62 VINTSKEMTSFSDFPPEDTMANFMHNTEMCRYLNNYAKHHELL 104
> 7292718
Length=685
Score = 29.6 bits (65), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 29/95 (30%), Positives = 39/95 (41%), Gaps = 3/95 (3%)
Query 3 PFLADLEPLGWIHTQPNENPQLLPRDVLAHAKILNENKNWDAATAAVVTCAFTPGSCSLT 62
P +AD L W+ QP+E QL R + A A+I + W A PG S
Sbjct 208 PLIADRCFLAWLVKQPSEQGQLRARQISA-AQINKLEELWKENIEATFQDLEKPGIDSEP 266
Query 63 AYRLT--PQGYQWGKANRDMSATPAGYSKGHAERA 95
A+ L GYQ+ K + A Y + E A
Sbjct 267 AHVLLRYEDGYQYEKTFGPLVRLEAEYDQKLKESA 301
> SPBC1271.15c
Length=686
Score = 28.5 bits (62), Expect = 7.0, Method: Composition-based stats.
Identities = 19/60 (31%), Positives = 23/60 (38%), Gaps = 1/60 (1%)
Query 90 GHAERAQMLLSDVFSGFFLVPAEGGGI-WNYNFMGVKFSPAMKYSLVLDTPKDFYHEAHR 148
GH + + L D F + E GGI V F K+ LDTP EA R
Sbjct 178 GHVDHGKTTLLDAFRKSTIASTEHGGITQKIGAFTVPFDKGSKFITFLDTPGHMAFEAMR 237
Lambda K H
0.319 0.134 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2598880752
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40