bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0767_orf2
Length=156
Score E
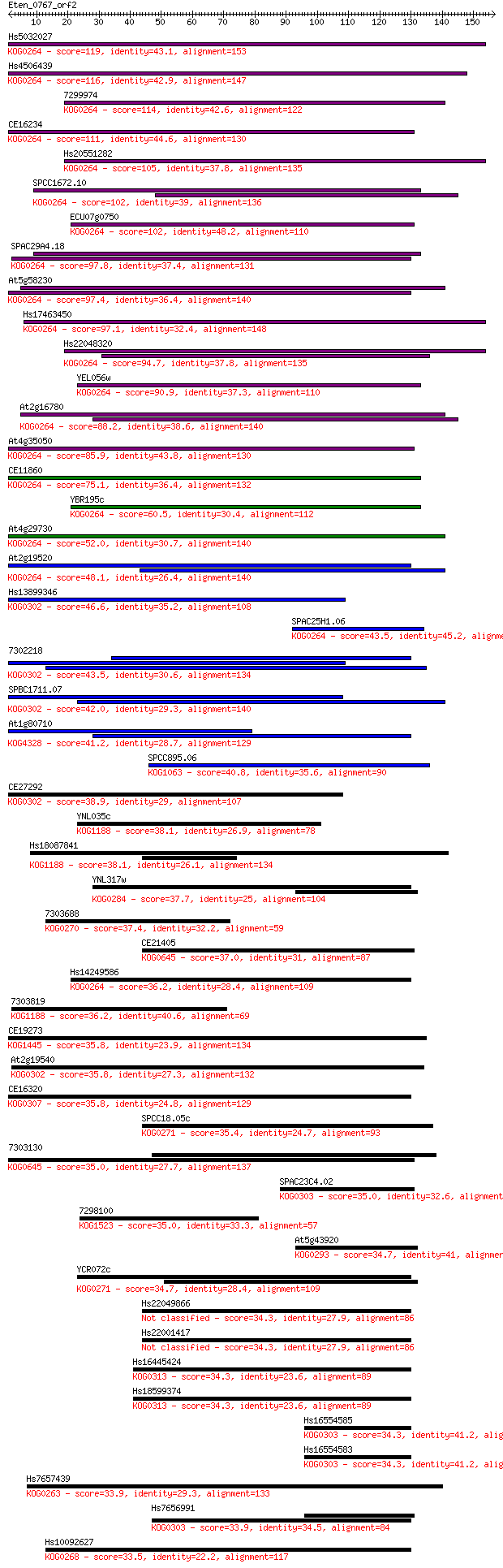
Sequences producing significant alignments: (Bits) Value
Hs5032027 119 4e-27
Hs4506439 116 2e-26
7299974 114 6e-26
CE16234 111 5e-25
Hs20551282 105 4e-23
SPCC1672.10 102 3e-22
ECU07g0750 102 3e-22
SPAC29A4.18 97.8 8e-21
At5g58230 97.4 1e-20
Hs17463450 97.1 1e-20
Hs22048320 94.7 7e-20
YEL056w 90.9 8e-19
At2g16780 88.2 6e-18
At4g35050 85.9 3e-17
CE11860 75.1 5e-14
YBR195c 60.5 1e-09
At4g29730 52.0 5e-07
At2g19520 48.1 6e-06
Hs13899346 46.6 2e-05
SPAC25H1.06 43.5 1e-04
7302218 43.5 2e-04
SPBC1711.07 42.0 4e-04
At1g80710 41.2 8e-04
SPCC895.06 40.8 0.001
CE27292 38.9 0.004
YNL035c 38.1 0.008
Hs18087841 38.1 0.008
YNL317w 37.7 0.010
7303688 37.4 0.011
CE21405 37.0 0.014
Hs14249586 36.2 0.024
7303819 36.2 0.025
CE19273 35.8 0.032
At2g19540 35.8 0.033
CE16320 35.8 0.035
SPCC18.05c 35.4 0.041
7303130 35.0 0.052
SPAC23C4.02 35.0 0.063
7298100 35.0 0.065
At5g43920 34.7 0.086
YCR072c 34.7 0.088
Hs22049866 34.3 0.096
Hs22001417 34.3 0.097
Hs16445424 34.3 0.10
Hs18599374 34.3 0.11
Hs16554585 34.3 0.11
Hs16554583 34.3 0.11
Hs7657439 33.9 0.13
Hs7656991 33.9 0.15
Hs10092627 33.5 0.17
> Hs5032027
Length=425
Score = 119 bits (297), Expect = 4e-27, Method: Compositional matrix adjust.
Identities = 66/183 (36%), Positives = 99/183 (54%), Gaps = 39/183 (21%)
Query 1 DDGQLNIWDLRAG--ASPVH----------------------------RGLSVWDFRDLR 30
DD +L IWD R+ + P H + +++WD R+L+
Sbjct 248 DDQKLMIWDTRSNNTSKPSHSVDAHTAEVNCLSFNPYSEFILATGSADKTVALWDLRNLK 307
Query 31 RPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEV 90
H +DE + ++W P N+ +LA+ TDR + ++D S IG EQSPE+AE+G PE+
Sbjct 308 LKLHSFESHKDE-IFQVQWSPHNETILASSGTDRRLNVWDLSKIGEEQSPEDAEDGPPEL 366
Query 91 IFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKTFEPEEANFLYEEGSM 150
+F+HGGH A I++ WNP E PW I S SEDN++Q+WQ + + E+ EGS+
Sbjct 367 LFIHGGHTAKISDFSWNPNE---PWVICSVSEDNIMQVWQMAENIYNDEDP-----EGSV 418
Query 151 DEE 153
D E
Sbjct 419 DPE 421
> Hs4506439
Length=425
Score = 116 bits (291), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 63/177 (35%), Positives = 93/177 (52%), Gaps = 34/177 (19%)
Query 1 DDGQLNIWDLRAGAS--PVH----------------------------RGLSVWDFRDLR 30
DD +L IWD R+ + P H + +++WD R+L+
Sbjct 247 DDQKLMIWDTRSNTTSKPSHLVDAHTAEVNCLSFNPYSEFILATGSADKTVALWDLRNLK 306
Query 31 RPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEV 90
H +DE + W P N+ +LA+ TDR + ++D S IG EQS E+AE+G PE+
Sbjct 307 LKLHTFESHKDEIF-QVHWSPHNETILASSGTDRRLNVWDLSKIGEEQSAEDAEDGPPEL 365
Query 91 IFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKTFEPEEANFLYEE 147
+F+HGGH A I++ WNP E PW I S SEDN++QIWQ + + EE++ E
Sbjct 366 LFIHGGHTAKISDFSWNPNE---PWVICSVSEDNIMQIWQMAENIYNDEESDVTTSE 419
> 7299974
Length=430
Score = 114 bits (286), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 52/122 (42%), Positives = 79/122 (64%), Gaps = 4/122 (3%)
Query 19 RGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQ 78
+ +++WD R+L+ H +DE ++W P N+ +LA+ TDR + ++D S IG EQ
Sbjct 300 KTVALWDLRNLKLKLHSFESHKDEIF-QVQWSPHNETILASSGTDRRLHVWDLSKIGEEQ 358
Query 79 SPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKTFEP 138
S E+AE+G PE++F+HGGH A I++ WNP E PW I S SEDN++Q+WQ + +
Sbjct 359 STEDAEDGPPELLFIHGGHTAKISDFSWNPNE---PWIICSVSEDNIMQVWQMAENVYND 415
Query 139 EE 140
EE
Sbjct 416 EE 417
> CE16234
Length=417
Score = 111 bits (278), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 58/158 (36%), Positives = 88/158 (55%), Gaps = 33/158 (20%)
Query 1 DDGQLNIWDLRAGASPVH----------------------------RGLSVWDFRDLRRP 32
DD +L IWD+R ++P H + +++WD R+LR
Sbjct 243 DDKKLLIWDVRT-STPGHCIDAHSAEVNCLAFNPYSEFILATGSADKTVALWDLRNLRMK 301
Query 33 AHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIF 92
H DE + ++W P N+ +LA+ TD+ + ++D S IG +QS E+AE+G PE++F
Sbjct 302 LHSFESHRDE-IFQVQWSPHNETILASSGTDKRLHVWDLSKIGEDQSAEDAEDGPPELLF 360
Query 93 VHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQ 130
+HGGH A I++ WNP E PW + S SEDN+LQ+WQ
Sbjct 361 IHGGHTAKISDFSWNPNE---PWVVCSVSEDNILQVWQ 395
> Hs20551282
Length=356
Score = 105 bits (262), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 51/135 (37%), Positives = 80/135 (59%), Gaps = 9/135 (6%)
Query 19 RGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQ 78
+ +++ D R+L+ H L + + +W P N+ +LA+ T+ + ++D S IG +Q
Sbjct 227 KTVALRDLRNLKLKLHSF-ELHKDKIFQFQWSPHNETILASSGTNHRLNVWDLSKIGEKQ 285
Query 79 SPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKTFEP 138
SPE+ E+G PE++F+HGGH A I + WNP E PW I S EDN++Q+WQ + +
Sbjct 286 SPEDKEDGPPELLFIHGGHTAKIPDFSWNPNE---PWVICSVPEDNIMQVWQMAENIYND 342
Query 139 EEANFLYEEGSMDEE 153
E Y EGS+D E
Sbjct 343 E-----YPEGSVDPE 352
> SPCC1672.10
Length=430
Score = 102 bits (255), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 51/124 (41%), Positives = 78/124 (62%), Gaps = 4/124 (3%)
Query 9 DLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRI 68
D + + +++WD R+ + H L EDE + L+W P ++ +LA+ +TDR V I
Sbjct 294 DYLLATASADKTVALWDLRNPYQRLHTLEGHEDE-VYGLEWSPHDEPILASSSTDRRVCI 352
Query 69 FDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQI 128
+D IG EQ+PE+AE+G+PE++F+HGGH I+E W P E W + S ++DN+LQI
Sbjct 353 WDLEKIGEEQTPEDAEDGSPELLFMHGGHTNRISEFSWCPNER---WVVGSLADDNILQI 409
Query 129 WQPS 132
W PS
Sbjct 410 WSPS 413
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/97 (23%), Positives = 40/97 (41%), Gaps = 9/97 (9%)
Query 48 KWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWN 107
++ PQ ++A IFD + A + E P+ + GH A + WN
Sbjct 138 RYMPQKPEIIATMGEGGNAYIFDTTCHDALTT----GEALPQAVL--KGHTAEGFGLCWN 191
Query 108 PIEDKFPWAIASASEDNVLQIWQPSVKTFEPEEANFL 144
P P +A+ +ED V+ +W ++F E +
Sbjct 192 P---NLPGNLATGAEDQVICLWDVQTQSFTSSETKVI 225
> ECU07g0750
Length=384
Score = 102 bits (254), Expect = 3e-22, Method: Compositional matrix adjust.
Identities = 53/110 (48%), Positives = 70/110 (63%), Gaps = 4/110 (3%)
Query 21 LSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSP 80
+ VWD R L +P H LL + + S++W P N VLA+G+TDR V ++D GAE
Sbjct 274 VKVWDRRSLSQPLHILLG-HSKDVVSVEWSPHNDKVLASGSTDRRVIVWDLGQAGAEVPE 332
Query 81 EEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQ 130
E EG PE+ F+HGGH + + +I WNP E P+ IAS SEDN+LQIWQ
Sbjct 333 EYKAEGPPEMKFLHGGHTSTVCDISWNPAE---PFEIASVSEDNILQIWQ 379
> SPAC29A4.18
Length=431
Score = 97.8 bits (242), Expect = 8e-21, Method: Compositional matrix adjust.
Identities = 46/124 (37%), Positives = 75/124 (60%), Gaps = 4/124 (3%)
Query 9 DLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRI 68
D + +++WD R+L + H L ED +T + + P + +LA+ + DR +
Sbjct 294 DFILATCSTDKTIALWDLRNLNQRLHTLEGHED-IVTKISFSPHEEPILASTSADRRTLV 352
Query 69 FDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQI 128
+D S IG +Q EEA++G PE++F+HGGH + ++DW P + W +A+A+EDN+LQI
Sbjct 353 WDLSRIGEDQPAEEAQDGPPELLFMHGGHTSCTIDMDWCP---NYNWTMATAAEDNILQI 409
Query 129 WQPS 132
W PS
Sbjct 410 WTPS 413
Score = 30.0 bits (66), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 26/128 (20%), Positives = 54/128 (42%), Gaps = 24/128 (18%)
Query 2 DGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGA 61
D L+ WDL A ++ D + ++ ++ +++H +++ +LA+ +
Sbjct 206 DATLSCWDLNA-----------YNESDSASVLKVHISSHEKQVSDVRFHYKHQDLLASVS 254
Query 62 TDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASAS 121
D+Y+ + D A P + VH H I + +NP D + +A+ S
Sbjct 255 YDQYLHVHDIRRPDASTKPARS---------VH-AHSGPIHSVAFNPHND---FILATCS 301
Query 122 EDNVLQIW 129
D + +W
Sbjct 302 TDKTIALW 309
> At5g58230
Length=424
Score = 97.4 bits (241), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 48/136 (35%), Positives = 81/136 (59%), Gaps = 6/136 (4%)
Query 5 LNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDR 64
N W + G++ + + ++D R L H ++E + W+P+N+ +LA+ R
Sbjct 284 FNEWVVATGST--DKTVKLFDLRKLSTALHTFDSHKEEVF-QVGWNPKNETILASCCLGR 340
Query 65 YVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDN 124
+ ++D S I EQ+ E+AE+G PE++F+HGGH + I++ WNP ED W I+S +EDN
Sbjct 341 RLMVWDLSRIDEEQTVEDAEDGPPELLFIHGGHTSKISDFSWNPCED---WVISSVAEDN 397
Query 125 VLQIWQPSVKTFEPEE 140
+LQIWQ + + E+
Sbjct 398 ILQIWQMAENIYHDED 413
Score = 32.0 bits (71), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 26/129 (20%), Positives = 58/129 (44%), Gaps = 26/129 (20%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
DD Q+ +WD+ A+P ++ L A ++ + + + WH +++ + +
Sbjct 199 DDAQICLWDI--NATPKNKSLD----------AQQIFKAHEGVVEDVAWHLRHEYLFGSV 246
Query 61 ATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASA 120
D+Y+ I+D +SP A + V+ H + + +NP + W +A+
Sbjct 247 GDDQYLLIWDL------RSP-SASKPVQSVV----AHSMEVNCLAFNPFNE---WVVATG 292
Query 121 SEDNVLQIW 129
S D ++++
Sbjct 293 STDKTVKLF 301
> Hs17463450
Length=396
Score = 97.1 bits (240), Expect = 1e-20, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 84/148 (56%), Gaps = 9/148 (6%)
Query 6 NIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRY 65
++++ + +++ D R+L+ H L+D+ ++W P N+ +LA+ T+
Sbjct 254 SLYEFILATGSADKTVALRDLRNLKLKLHSFELLKDKIF-QVQWSPHNETILASSGTNHR 312
Query 66 VRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNV 125
+ ++D S IG +QSPE+ ++ PE++F+HGGH A I + NP E PW I S EDN+
Sbjct 313 LNVWDLSKIGEKQSPEDKKDRPPELLFIHGGHTAKIPDFSGNPNE---PWVICSVPEDNI 369
Query 126 LQIWQPSVKTFEPEEANFLYEEGSMDEE 153
+Q+WQ + + E+ EGS+D E
Sbjct 370 MQVWQMAENIYNNEDP-----EGSVDPE 392
> Hs22048320
Length=345
Score = 94.7 bits (234), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 51/135 (37%), Positives = 80/135 (59%), Gaps = 10/135 (7%)
Query 19 RGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQ 78
+ +++WD R+L+ H +DE + ++W P N+ +LA+ TD + ++D S I EQ
Sbjct 210 KTVALWDLRNLKLKVHSFESHKDE-IFQVQWSPHNETILASSGTDHGLNVWDLSKITEEQ 268
Query 79 SPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKTFEP 138
SPE+AE+ PE +F+HGGH A I + NP E PW I S SE+N++++WQ +
Sbjct 269 SPEDAEDRPPE-LFIHGGHTAKIPDFSLNPSE---PWMICSVSENNIMEVWQMAENISNG 324
Query 139 EEANFLYEEGSMDEE 153
E+ EGS+D E
Sbjct 325 EDP-----EGSVDPE 334
Score = 30.4 bits (67), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 26/105 (24%), Positives = 42/105 (40%), Gaps = 12/105 (11%)
Query 31 RPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEV 90
P RL + E L W+P L + + D V ++D S I +EG +
Sbjct 83 NPDLRLCGHQKEGY-GLSWNPNLSGHLLSASDDHTVCLWDISAI--------PKEGKVDA 133
Query 91 IFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKT 135
+ GH A + ++ WN + + +A +D L IW K
Sbjct 134 RTIFTGHTAVVEDVFWNLLHESLFRLVA---DDQKLMIWDTRSKN 175
> YEL056w
Length=401
Score = 90.9 bits (224), Expect = 8e-19, Method: Compositional matrix adjust.
Identities = 41/110 (37%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query 23 VWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEE 82
++D R+++ P H + ED A+ +L++ V+ + +D + ++D IGAEQ+P++
Sbjct 278 LYDLRNMKEPLHHMSGHED-AVNNLEFSTHVDGVVVSSGSDNRLMMWDLKQIGAEQTPDD 336
Query 83 AEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPS 132
AE+G PE+I VH GH + + + D NP + PW +ASA E+N+LQ+W+ S
Sbjct 337 AEDGVPELIMVHAGHRSSVNDFDLNP---QIPWLVASAEEENILQVWKCS 383
> At2g16780
Length=415
Score = 88.2 bits (217), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 52/138 (37%), Positives = 77/138 (55%), Gaps = 8/138 (5%)
Query 5 LNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDR 64
N W L +S ++++D R L P H + E E ++W P ++ VLA+ DR
Sbjct 271 FNEWVLATASS--DSTVALFDLRKLNAPLHVMSSHEGEVF-QVEWDPNHETVLASSGEDR 327
Query 65 YVRIFDCSLIGAEQSPEE--AEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASE 122
+ ++D + +G EQ E AE+G PE++F HGGH A I++ WN E PW IAS +E
Sbjct 328 RLMVWDLNRVGEEQLEIELDAEDGPPELLFSHGGHKAKISDFAWNKNE---PWVIASVAE 384
Query 123 DNVLQIWQPSVKTFEPEE 140
DN LQ+WQ + + EE
Sbjct 385 DNSLQVWQMAESIYRDEE 402
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 35/124 (28%), Positives = 54/124 (43%), Gaps = 25/124 (20%)
Query 28 DLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGA 87
DLR H D+ L W P + L +G+ D+ + ++D S +P++ A
Sbjct 161 DLRLVGH------DKEGYGLSWSPFKEGYLLSGSQDQKICLWDVS-----ATPQDKVLNA 209
Query 88 PEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW-------QPSVKTFEPEE 140
+FV+ GH + I ++ W+ + SA ED L IW Q VK E E
Sbjct 210 ---MFVYEGHESAIADVSWHMKNENL---FGSAGEDGRLVIWDTRTNQMQHQVKVHE-RE 262
Query 141 ANFL 144
N+L
Sbjct 263 VNYL 266
> At4g35050
Length=424
Score = 85.9 bits (211), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 57/159 (35%), Positives = 79/159 (49%), Gaps = 33/159 (20%)
Query 1 DDGQLNIWDLRAGAS----PVHR-----------------------GLSVWDFRDLRRPA 33
DD QL IWDLR VH ++++D R L P
Sbjct 239 DDCQLVIWDLRTNQMQHQVKVHEREINYLSFNPFNEWVLATASSDSTVALFDLRKLTAPL 298
Query 34 HRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEE--AEEGAPEVI 91
H L E E ++W P ++ VLA+ DR + ++D + +G EQ E AE+G PE++
Sbjct 299 HVLSKHEGEVF-QVEWDPNHETVLASSGEDRRLMVWDINRVGDEQLEIELDAEDGPPELL 357
Query 92 FVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQ 130
F HGGH A I++ WN E PW I+S +EDN LQ+WQ
Sbjct 358 FSHGGHKAKISDFAWNKDE---PWVISSVAEDNSLQVWQ 393
> CE11860
Length=412
Score = 75.1 bits (183), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 48/164 (29%), Positives = 74/164 (45%), Gaps = 39/164 (23%)
Query 1 DDGQLNIWDLRA---------------------------GASPVHRGLSVWDFRDLRRPA 33
DD +LN+WDLR V + +++WD R++R+
Sbjct 242 DDRKLNLWDLRQSKPQLTAVGHTAEVNCITFNPFSEYILATGSVDKTVALWDMRNMRKKM 301
Query 34 HRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGA-----P 88
+ L + DE + + P + VLA+ +D V ++D S I Q P + + P
Sbjct 302 YTLKHHNDEIF-QVSFSPHYETVLASSGSDDRVIVWDISKI---QDPSSSSAASSDSVPP 357
Query 89 EVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPS 132
EVIF+H GH + + WNP PW I S+ E N LQ+W+ S
Sbjct 358 EVIFIHAGHTGKVADFSWNP---NRPWTICSSDEFNALQVWEVS 398
> YBR195c
Length=422
Score = 60.5 bits (145), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/113 (30%), Positives = 58/113 (51%), Gaps = 19/113 (16%)
Query 21 LSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLA-AGATDRYVRIFDCSLIGAEQS 79
L++WD R++ + + ++++L+W P VLA AG D V+++D S
Sbjct 321 LNLWDIRNMNKSPIATME-HGTSVSTLEWSPNFDTVLATAGQEDGLVKLWDTS------- 372
Query 80 PEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPS 132
E IF HGGH+ G+ +I W+ + PW + S + DN + IW+P+
Sbjct 373 -------CEETIFTHGGHMLGVNDISWDAHD---PWLMCSVANDNSVHIWKPA 415
> At4g29730
Length=496
Score = 52.0 bits (123), Expect = 5e-07, Method: Composition-based stats.
Identities = 43/181 (23%), Positives = 70/181 (38%), Gaps = 47/181 (25%)
Query 1 DDGQLNIWDLRAGASPVHR-----------------------------GLSVWDFRDLRR 31
DD L +WD R G SP + + V+D R+L
Sbjct 302 DDSCLMLWDARTGTSPAMKVEKAHDADLHCVDWNPHDNNLILTGSADNTVRVFDRRNLTS 361
Query 32 -----PAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEG 86
P ++ A+ ++W P +V + A D + I+DC +G + E A +
Sbjct 362 NGVGSPVYKF-EGHRAAVLCVQWSPDKSSVFGSSAEDGLLNIWDCDRVG--KKSERATKT 418
Query 87 APEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASED-------NVLQIWQPSVKTFEPE 139
+ F H GH + + W+ + PW I S S++ LQIW+ S + PE
Sbjct 419 PDGLFFQHAGHRDKVVDFHWSLLN---PWTIVSVSDNCESIGGGGTLQIWRMSDLIYRPE 475
Query 140 E 140
+
Sbjct 476 D 476
> At2g19520
Length=507
Score = 48.1 bits (113), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 33/129 (25%), Positives = 53/129 (41%), Gaps = 27/129 (20%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
DD L +WD R G +PV ++ D L + W+P + ++ G
Sbjct 313 DDSCLILWDARTGTNPV----------------TKVEKAHDADLHCVDWNPHDDNLILTG 356
Query 61 ATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASA 120
+ D VR+FD + A G I+ GH A + + W+P + S+
Sbjct 357 SADNTVRLFDRRKLTA--------NGVGSPIYKFEGHKAAVLCVQWSPDKSSV---FGSS 405
Query 121 SEDNVLQIW 129
+ED +L IW
Sbjct 406 AEDGLLNIW 414
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 29/105 (27%), Positives = 47/105 (44%), Gaps = 12/105 (11%)
Query 43 ALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGIT 102
A+ ++W P +V + A D + I+D + + + A + + F H GH +
Sbjct 388 AVLCVQWSPDKSSVFGSSAEDGLLNIWDYDRVSKKS--DRAAKSPAGLFFQHAGHRDKVV 445
Query 103 EIDWNPIEDKFPWAIASASED-------NVLQIWQPSVKTFEPEE 140
+ WN + PW I S S+D LQIW+ S + PEE
Sbjct 446 DFHWNASD---PWTIVSVSDDCETTGGGGTLQIWRMSDLIYRPEE 487
> Hs13899346
Length=446
Score = 46.6 bits (109), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 38/116 (32%), Positives = 54/116 (46%), Gaps = 27/116 (23%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
DDG L IWDLR S G V F+ P +TS++WHPQ+ V AA
Sbjct 328 DDGALKIWDLRQFKS----GSPVATFKQHVAP-----------VTSVEWHPQDSGVFAAS 372
Query 61 ATDRYVRIFDCSLIGAEQSPE----EAEEGAP----EVIFVHGGHVAGITEIDWNP 108
D + +D + E+ PE EA+ G +++FVH G + E+ W+P
Sbjct 373 GADHQITQWD---LAVERDPEAGDVEADPGLADLPQQLLFVHQGETE-LKELHWHP 424
> SPAC25H1.06
Length=408
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 28/42 (66%), Gaps = 3/42 (7%)
Query 92 FVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSV 133
FVH GH + E+D++P E + IAS ++DN L IW+P+V
Sbjct 367 FVHAGHKGTVNEVDFDPFEAQ---CIASVADDNELHIWKPNV 405
> 7302218
Length=422
Score = 43.5 bits (101), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 48/96 (50%), Gaps = 14/96 (14%)
Query 34 HRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFV 93
R L +++ L+W P ++VLA+ + D+ +RI+DC SP++A ++
Sbjct 228 QRPLAGHSQSVEDLQWSPNERSVLASCSVDKTIRIWDC-----RASPQKA-----CMLTC 277
Query 94 HGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
H + + I WN E P+ IAS +D L IW
Sbjct 278 EDAHQSDVNVISWNRNE---PF-IASGGDDGYLHIW 309
Score = 43.1 bits (100), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 54/115 (46%), Gaps = 23/115 (20%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
DDG L+IWDLR S ++P + D +T+++W P VLA+G
Sbjct 302 DDGYLHIWDLRQFQS--------------KKPIATFKHHTDH-ITTVEWSPAEATVLASG 346
Query 61 ATDRYVRIFDCSL-------IGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNP 108
D + ++D ++ + Q+ + + P+++F+H G I E+ W+P
Sbjct 347 GDDDQIALWDLAVEKDIDQAVDPAQNEDVLNKLPPQLLFIHQGQ-KEIKELHWHP 400
Score = 35.0 bits (79), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 30/125 (24%), Positives = 57/125 (45%), Gaps = 19/125 (15%)
Query 13 GASPVHRGLSVWDFRDLRRPAHRLLYLED---EALTSLKWHPQNKAVLAAGATDRYVRIF 69
+ V + + +WD R + A +L ED + + W+ +N+ +A+G D Y+ I+
Sbjct 252 ASCSVDKTIRIWDCRASPQKAC-MLTCEDAHQSDVNVISWN-RNEPFIASGGDDGYLHIW 309
Query 70 DCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
D + + P F H H IT ++W+P E +AS +D+ + +W
Sbjct 310 DL---------RQFQSKKPIATFKH--HTDHITTVEWSPAEATV---LASGGDDDQIALW 355
Query 130 QPSVK 134
+V+
Sbjct 356 DLAVE 360
> SPBC1711.07
Length=480
Score = 42.0 bits (97), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 30/112 (26%), Positives = 52/112 (46%), Gaps = 17/112 (15%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
D+G ++WDLR+ S V F+ R P + S++WHP +V+
Sbjct 358 DNGVWSVWDLRSLKSSSSVATPVASFKWHRAPIY-----------SIEWHPNEDSVIGVV 406
Query 61 ATDRYVRIFDCSL-IGAEQSPEEAEEG----APEVIFVHGGHVAGITEIDWN 107
D + ++D S+ + E+ A EG P+++F+H G I E+ W+
Sbjct 407 GADNQISLWDLSVELDEEEQDSRAAEGLQDVPPQLMFIHMGQ-QEIKEMHWH 457
Score = 30.0 bits (66), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 29/119 (24%), Positives = 50/119 (42%), Gaps = 11/119 (9%)
Query 23 VWDFRDLRRPAHRLLYLEDEA-LTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPE 81
+WD R+ ++ + + + L W+ + +LA GA + ++D + + S
Sbjct 318 IWDVRNKQKTSALTVNAHPGVDVNVLSWNTRVPNLLATGADNGVWSVWDLRSLKSSSSV- 376
Query 82 EAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKTFEPEE 140
P F H A I I+W+P ED I DN + +W SV+ E E+
Sbjct 377 ----ATPVASF--KWHRAPIYSIEWHPNEDS---VIGVVGADNQISLWDLSVELDEEEQ 426
> At1g80710
Length=516
Score = 41.2 bits (95), Expect = 8e-04, Method: Composition-based stats.
Identities = 24/78 (30%), Positives = 38/78 (48%), Gaps = 18/78 (23%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
D G N+WDLRAG S H W+ + R + S+ ++PQN V+A
Sbjct 326 DYGVFNVWDLRAGKSVFH-----WELHERR-------------INSIDFNPQNPHVMATS 367
Query 61 ATDRYVRIFDCSLIGAEQ 78
+TD ++D +GA++
Sbjct 368 STDGTACLWDLRSMGAKK 385
Score = 28.5 bits (62), Expect = 4.9, Method: Composition-based stats.
Identities = 25/102 (24%), Positives = 41/102 (40%), Gaps = 17/102 (16%)
Query 28 DLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGA 87
D+ + L+Y DEA+ SL P ++ L G ++D A
Sbjct 292 DVEKSVFDLVYSTDEAIFSLSQRPNDEQSLYFGQDYGVFNVWDLR--------------A 337
Query 88 PEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
+ +F H I ID+NP + P +A++S D +W
Sbjct 338 GKSVFHWELHERRINSIDFNP---QNPHVMATSSTDGTACLW 376
> SPCC895.06
Length=760
Score = 40.8 bits (94), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 32/108 (29%), Positives = 51/108 (47%), Gaps = 22/108 (20%)
Query 46 SLKWHPQNKAVLAAGATDRYVRIFDCSLIGA--EQSPEEAEEGA---PEVIFVHG----- 95
S K + A LA+G+ DRY+R+++ SL G+ E+ EE E V F G
Sbjct 198 SFKKTSGSTATLASGSQDRYIRLWNISLWGSEDEKVSEEFFESVLSNKPVRFTLGKIDLK 257
Query 96 --------GHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQPSVKT 135
GH + +DW+P ++ I S+S D+ + +W+P T
Sbjct 258 IVFDALLMGHEDWVMSVDWHPTKE----MILSSSADSSMIVWEPDTNT 301
> CE27292
Length=453
Score = 38.9 bits (89), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 31/108 (28%), Positives = 45/108 (41%), Gaps = 20/108 (18%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
DDG+L IW L+ + G V F+ P +TS+ WHP A
Sbjct 342 DDGELKIWSLKT----IQFGQPVALFKYHNSP-----------ITSVDWHPHETTTFMAS 386
Query 61 ATDRYVRIFDCSLIGAEQSPEEAEEGA-PEVIFVHGGHVAGITEIDWN 107
D I+D I E + EG P+++FVH G + E+ W+
Sbjct 387 GEDDQTTIWD---IATEADGQTNIEGVPPQLMFVHMGQNE-VKEVHWH 430
> YNL035c
Length=389
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 21/78 (26%), Positives = 36/78 (46%), Gaps = 7/78 (8%)
Query 23 VWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEE 82
++D R P L+ + +T +K+HP + +L +G+TD Y I+D +E
Sbjct 130 IYDIRKWDTPLRSLIDSHHDDVTCIKFHPSDVNILLSGSTDGYTNIYDL-------KQDE 182
Query 83 AEEGAPEVIFVHGGHVAG 100
E+ +VI H G
Sbjct 183 EEDALHQVINYASIHSCG 200
> Hs18087841
Length=387
Score = 38.1 bits (87), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 35/142 (24%), Positives = 64/142 (45%), Gaps = 31/142 (21%)
Query 8 WDLRAGAS--PVHRGLSVWDFRDLRRPAHRLLYLEDEALTSL---KWHPQNKAVLAAGAT 62
WDL + PV R L++ D R++ + ++++AL L +H + + G T
Sbjct 244 WDLNHLDTDEPVTR-LNIQDVREV-------VNMKEDALDYLIGGLYHEKTDTLHVIGGT 295
Query 63 DR-YVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASAS 121
++ + + +CS+ G V + GGH A + WN +D ++ +
Sbjct 296 NKGRIHLMNCSM-----------SGLTHVTSLQGGHAATVRSFCWNVQDD----SLLTGG 340
Query 122 EDNVLQIWQPSV--KTFEPEEA 141
ED L +W+P KTF +E+
Sbjct 341 EDAQLLLWKPGAIEKTFTKKES 362
Score = 28.9 bits (63), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 9/30 (30%), Positives = 21/30 (70%), Gaps = 0/30 (0%)
Query 44 LTSLKWHPQNKAVLAAGATDRYVRIFDCSL 73
+T +++HP N ++ +G++D V +FD ++
Sbjct 173 VTQVRFHPSNPNMVVSGSSDGLVNVFDINI 202
> YNL317w
Length=465
Score = 37.7 bits (86), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 25/103 (24%), Positives = 47/103 (45%), Gaps = 16/103 (15%)
Query 28 DLRRPAHRLLYLEDEA-LTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEG 86
D+R L+ + DE +L+WHP N+++ D ++ FD L+ P
Sbjct 290 DIRYSMKELMCVRDETDYMTLEWHPINESMFTLACYDGSLKHFD--LLQNLNEP------ 341
Query 87 APEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
++ + H IT + +NP+ F A+A++D ++ W
Sbjct 342 ---ILTIPYAHDKCITSLSYNPVGHIF----ATAAKDRTIRFW 377
Score = 28.9 bits (63), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/39 (38%), Positives = 22/39 (56%), Gaps = 4/39 (10%)
Query 93 VHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQP 131
V GH + DW+P IASAS+DN++++W P
Sbjct 214 VLSGHHWDVKSCDWHPEMG----LIASASKDNLVKLWDP 248
> 7303688
Length=459
Score = 37.4 bits (85), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 19/59 (32%), Positives = 31/59 (52%), Gaps = 2/59 (3%)
Query 13 GASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDC 71
+ V + + +WD D +P H + + + SL++HPQ + G D YVR+FDC
Sbjct 251 ASGSVDQTVILWDM-DEGQP-HTTITAFGKQIQSLEFHPQEAQSILTGCADGYVRLFDC 307
> CE21405
Length=314
Score = 37.0 bits (84), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 27/87 (31%), Positives = 42/87 (48%), Gaps = 10/87 (11%)
Query 44 LTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITE 103
L S+ W+ N V+A G D +R+F S +PE E + V G H +
Sbjct 235 LYSVAWNSTND-VIATGGGDCKIRLFKIS-----STPESP---VIEHLGVVGRHELDVNH 285
Query 104 IDWNPIEDKFPWAIASASEDNVLQIWQ 130
+ WNP KF + SAS+D +++W+
Sbjct 286 VAWNP-NPKFSNLLTSASDDGTIRLWE 311
> Hs14249586
Length=378
Score = 36.2 bits (82), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 49/124 (39%), Gaps = 19/124 (15%)
Query 21 LSVWDFRDLRRPAHRL-----LYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIG 75
L + D RDL P + + D L + W P K LA D V+++D +
Sbjct 250 LCLLDPRDLCHPVSSVQCPVSVPSPDPELLRVTWAPGLKNCLAISGFDGTVQVYDATSWD 309
Query 76 AEQSPEEAEEGAPEVIFVHGGHV----------AGITEIDWNPIEDKFPWAIASASEDNV 125
+S ++ E +F H GH+ +T W+P P + SA+ D
Sbjct 310 GTRS-QDGTRSQVEPLFTHRGHIFLDGNGMDPAPLVTTHTWHPCR---PRTLLSATNDAS 365
Query 126 LQIW 129
L +W
Sbjct 366 LHVW 369
> 7303819
Length=411
Score = 36.2 bits (82), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 28/106 (26%), Positives = 43/106 (40%), Gaps = 37/106 (34%)
Query 2 DGQLNIWDLRA-------------GASPVHRGLSVWD----------------------F 26
DG + ++DLR PV + LS +D F
Sbjct 128 DGYVRLYDLRLRGEQARFKYTQHPNVPPVPKSLSCFDRNANGRIICCGTEQFHSNAFLVF 187
Query 27 RDLRRPAHRLLYLE--DEALTSLKWHPQNKAVLAAGATDRYVRIFD 70
D+R +Y E ++ +TSL++H QN +LA G+ D V +FD
Sbjct 188 FDVRERQQMGVYFESHEDDITSLRFHAQNPDLLATGSVDGLVNVFD 233
> CE19273
Length=1057
Score = 35.8 bits (81), Expect = 0.032, Method: Composition-based stats.
Identities = 32/134 (23%), Positives = 56/134 (41%), Gaps = 29/134 (21%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
D GQ+N+W L P R+ P +++ + E +TSL+WHP +LA
Sbjct 649 DCGQINLWRLTTNDGP----------RNEMEP-EKIIKIGGEKITSLRWHPLASDLLAVA 697
Query 61 ATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASA 120
++ + ++D + FV+ H GI I W+ + IAS
Sbjct 698 LSNSTIELWDVA------------NAKLYSRFVN--HTGGILGIAWSADGRR----IASV 739
Query 121 SEDNVLQIWQPSVK 134
+D L + +P+ +
Sbjct 740 GKDATLFVHEPASR 753
> At2g19540
Length=469
Score = 35.8 bits (81), Expect = 0.033, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 52/132 (39%), Gaps = 30/132 (22%)
Query 2 DGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGA 61
DG + +WD+R G SP L AH + + + W+ +LA+G+
Sbjct 291 DGSVAVWDIRLGKSPA-----------LSFKAH------NADVNVISWNRLASCMLASGS 333
Query 62 TDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASAS 121
D I D LI +G V+ H IT I+W+ E +A S
Sbjct 334 DDGTFSIRDLRLI----------KGGDAVVAHFEYHKHPITSIEWSAHEAS---TLAVTS 380
Query 122 EDNVLQIWQPSV 133
DN L IW S+
Sbjct 381 GDNQLTIWDLSL 392
> CE16320
Length=1083
Score = 35.8 bits (81), Expect = 0.035, Method: Compositional matrix adjust.
Identities = 32/132 (24%), Positives = 53/132 (40%), Gaps = 16/132 (12%)
Query 1 DDGQLNIWDLR---AGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVL 57
D +L W+L+ AS R +S WD R P + +SL W+P + + L
Sbjct 166 DQVKLLRWNLKNESVFASISSRRVSFWDLRRNGSPVLEFAEIPGCDWSSLSWNPSDASQL 225
Query 58 AAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAI 117
+ ++ + Q + P + H H GIT +DWN +D+ +
Sbjct 226 IVSSQSQHASVI--------QKWDSRFTSTPVKEYRH--HNMGITSVDWNKADDRL---V 272
Query 118 ASASEDNVLQIW 129
S+ D + IW
Sbjct 273 ISSGCDGQVVIW 284
> SPCC18.05c
Length=502
Score = 35.4 bits (80), Expect = 0.041, Method: Composition-based stats.
Identities = 23/96 (23%), Positives = 46/96 (47%), Gaps = 17/96 (17%)
Query 44 LTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITE 103
++ + W P + +++A G+ D +R +D ++G+P + H I
Sbjct 180 VSCVAWAP-DASIIATGSMDNTIRFWD------------PKKGSP-IGDALRRHTKPIMA 225
Query 104 IDWNPIE---DKFPWAIASASEDNVLQIWQPSVKTF 136
+ W P+ D P+ +AS S+DN ++IW ++T
Sbjct 226 LCWQPLHLAPDSGPYLLASGSKDNTVRIWNVKLRTL 261
> 7303130
Length=335
Score = 35.0 bits (79), Expect = 0.052, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 39/91 (42%), Gaps = 16/91 (17%)
Query 47 LKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDW 106
+ WHP+ V A+ D+ +RI+ SL G S + + GH I EI W
Sbjct 20 VAWHPKGN-VFASCGEDKAIRIW--SLTGNTWSTK---------TILSDGHKRTIREIRW 67
Query 107 NPIEDKFPWAIASASEDNVLQIWQPSVKTFE 137
+P +ASAS D IW S FE
Sbjct 68 SPCGQY----LASASFDATTAIWSKSSGEFE 94
Score = 29.3 bits (64), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 30/131 (22%), Positives = 53/131 (40%), Gaps = 13/131 (9%)
Query 1 DDGQLNIWDLRAGASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAG 60
DD + IW RA G++ D + + + + A+ + W + ++A
Sbjct 214 DDTTIKIW--RAYHPGNTAGVATPDQQTVWKCVCTVSGQHSRAIYDVSW-CKLTGLIATA 270
Query 61 ATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVH-GGHVAGITEIDWNPIEDKFPWAIAS 119
D +RIF E S + +E E I G H + + WNP+ + S
Sbjct 271 CGDDGIRIF------KESSDSKPDEPTFEQITAEEGAHDQDVNSVQWNPV---VAGQLIS 321
Query 120 ASEDNVLQIWQ 130
S+D ++IW+
Sbjct 322 CSDDGTIKIWK 332
> SPAC23C4.02
Length=601
Score = 35.0 bits (79), Expect = 0.063, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 25/43 (58%), Gaps = 3/43 (6%)
Query 88 PEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQ 130
P+ + + GH A + + DWNP D+ +AS +D+ + IW+
Sbjct 71 PDQVNLFRGHTAAVLDTDWNPFHDQ---VLASGGDDSKIMIWK 110
> 7298100
Length=377
Score = 35.0 bits (79), Expect = 0.065, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 29/57 (50%), Gaps = 8/57 (14%)
Query 24 WDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSP 80
W + +++P +TSL WHP N +L AG+TD VR+F + E+ P
Sbjct 136 WVSKHIKKPIR-------STVTSLDWHPNN-VLLLAGSTDYKVRVFSAFIKDIEEPP 184
> At5g43920
Length=523
Score = 34.7 bits (78), Expect = 0.086, Method: Composition-based stats.
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 3/39 (7%)
Query 93 VHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQP 131
V GH + + WNP K P +ASAS+D ++IW P
Sbjct 480 VLSGHSMTVNCVSWNP---KNPRMLASASDDQTIRIWGP 515
> YCR072c
Length=515
Score = 34.7 bits (78), Expect = 0.088, Method: Composition-based stats.
Identities = 30/110 (27%), Positives = 50/110 (45%), Gaps = 19/110 (17%)
Query 23 VWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEE 82
+WD D + P H L + L + W P + V+A G+ D +R++D P+
Sbjct 170 IWDC-DTQTPMHTLKGHYNWVLC-VSWSPDGE-VIATGSMDNTIRLWD---------PKS 217
Query 83 AEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWA---IASASEDNVLQIW 129
+ + GH IT + W PI P + +AS+S+D ++IW
Sbjct 218 GQCLGDAL----RGHSKWITSLSWEPIHLVKPGSKPRLASSSKDGTIKIW 263
Score = 33.5 bits (75), Expect = 0.18, Method: Composition-based stats.
Identities = 21/81 (25%), Positives = 34/81 (41%), Gaps = 18/81 (22%)
Query 51 PQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIE 110
P + + GA D RI+DC Q+P +G H + + W+P
Sbjct 153 PHTSSRMVTGAGDNTARIWDCD----TQTPMHTLKG----------HYNWVLCVSWSPDG 198
Query 111 DKFPWAIASASEDNVLQIWQP 131
+ IA+ S DN +++W P
Sbjct 199 E----VIATGSMDNTIRLWDP 215
> Hs22049866
Length=1106
Score = 34.3 bits (77), Expect = 0.096, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 11/92 (11%)
Query 44 LTSLKWH------PQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGH 97
+ ++ WH P+ ++A+G+ + + + + + E SPE + E GH
Sbjct 179 VNTISWHHEHGSQPELSYLMASGSNNAVIYVHNLKTV-IESSPE-SPVTITEPYRTLSGH 236
Query 98 VAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
A IT + W+P D + SAS D Q+W
Sbjct 237 TAKITSVAWSPHHDG---RLVSASYDGTAQVW 265
> Hs22001417
Length=1508
Score = 34.3 bits (77), Expect = 0.097, Method: Compositional matrix adjust.
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 11/92 (11%)
Query 44 LTSLKWH------PQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGH 97
+ ++ WH P+ ++A+G+ + + + + + E SPE + E GH
Sbjct 581 VNTISWHHEHGSQPELSYLMASGSNNAVIYVHNLKTV-IESSPE-SPVTITEPYRTLSGH 638
Query 98 VAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
A IT + W+P D + SAS D Q+W
Sbjct 639 TAKITSVAWSPHHDG---RLVSASYDGTAQVW 667
> Hs16445424
Length=423
Score = 34.3 bits (77), Expect = 0.10, Method: Compositional matrix adjust.
Identities = 21/89 (23%), Positives = 44/89 (49%), Gaps = 15/89 (16%)
Query 41 DEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAG 100
++ + + P K LA+G+TDR++R++D ++G+ V H
Sbjct 297 NKVFNCISYSPLCKR-LASGSTDRHIRLWD----------PRTKDGSL-VSLSLTSHTGW 344
Query 101 ITEIDWNPIEDKFPWAIASASEDNVLQIW 129
+T + W+P ++ + S S DN++++W
Sbjct 345 VTSVKWSPTHEQ---QLISGSLDNIVKLW 370
> Hs18599374
Length=423
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 21/89 (23%), Positives = 44/89 (49%), Gaps = 15/89 (16%)
Query 41 DEALTSLKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAG 100
++ + + P K LA+G+TDR++R++D ++G+ V H
Sbjct 297 NKVFNCISYSPLCKR-LASGSTDRHIRLWD----------PRTKDGSL-VSLSLTSHTGW 344
Query 101 ITEIDWNPIEDKFPWAIASASEDNVLQIW 129
+T + W+P ++ + S S DN++++W
Sbjct 345 VTSVKWSPTHEQ---QLISGSLDNIVKLW 370
> Hs16554585
Length=525
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 3/34 (8%)
Query 96 GHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
GH + ++ WNP +D + IAS SED ++IW
Sbjct 80 GHRGNVLDVKWNPFDD---FEIASCSEDATIKIW 110
> Hs16554583
Length=525
Score = 34.3 bits (77), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 14/34 (41%), Positives = 21/34 (61%), Gaps = 3/34 (8%)
Query 96 GHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
GH + ++ WNP +D + IAS SED ++IW
Sbjct 80 GHRGNVLDVKWNPFDD---FEIASCSEDATIKIW 110
> Hs7657439
Length=589
Score = 33.9 bits (76), Expect = 0.13, Method: Composition-based stats.
Identities = 39/140 (27%), Positives = 61/140 (43%), Gaps = 28/140 (20%)
Query 7 IWDLRAG------ASPVH-RGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAA 59
+WDL AS H R +W F D P R+ + +K+HP N LA
Sbjct 387 VWDLDISPYSLYFASGSHDRTARLWSF-DRTYPL-RIYAGHLADVDCVKFHP-NSNYLAT 443
Query 60 GATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIAS 119
G+TD+ VR++ A++G +F GH + + ++P K+ +AS
Sbjct 444 GSTDKTVRLW------------SAQQGNSVRLFT--GHRGPVLSLAFSP-NGKY---LAS 485
Query 120 ASEDNVLQIWQPSVKTFEPE 139
A ED L++W + T E
Sbjct 486 AGEDQRLKLWDLASGTLYKE 505
> Hs7656991
Length=474
Score = 33.9 bits (76), Expect = 0.15, Method: Composition-based stats.
Identities = 15/35 (42%), Positives = 20/35 (57%), Gaps = 3/35 (8%)
Query 96 GHVAGITEIDWNPIEDKFPWAIASASEDNVLQIWQ 130
GH + +IDW P D+ IAS SED + +WQ
Sbjct 78 GHTGPVLDIDWCPHNDQ---VIASGSEDCTVMVWQ 109
Score = 29.6 bits (65), Expect = 2.5, Method: Composition-based stats.
Identities = 24/83 (28%), Positives = 35/83 (42%), Gaps = 10/83 (12%)
Query 47 LKWHPQNKAVLAAGATDRYVRIFDCSLIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDW 106
+ W P N V+A+G+ D V ++ G S E P VI GH + + W
Sbjct 86 IDWCPHNDQVIASGSEDCTVMVWQIPENGLTLSLTE-----PVVIL--EGHSKRVGIVAW 138
Query 107 NPIEDKFPWAIASASEDNVLQIW 129
+P + SA DN + IW
Sbjct 139 HPTARN---VLLSAGCDNAIIIW 158
> Hs10092627
Length=445
Score = 33.5 bits (75), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 26/117 (22%), Positives = 51/117 (43%), Gaps = 20/117 (17%)
Query 13 GASPVHRGLSVWDFRDLRRPAHRLLYLEDEALTSLKWHPQNKAVLAAGATDRYVRIFDCS 72
G+ R + ++D R P +++ D ++ W+P + A D + FD
Sbjct 213 GSCASDRNIVLYDMRQAT-PLKKVIL--DMRTNTICWNPMEAFIFTAANEDYNLYTFD-- 267
Query 73 LIGAEQSPEEAEEGAPEVIFVHGGHVAGITEIDWNPIEDKFPWAIASASEDNVLQIW 129
+ A +P + VH HV+ + ++D++P +F SAS D ++I+
Sbjct 268 -MRALDTP----------VMVHMDHVSAVLDVDYSPTGKEF----VSASFDKSIRIF 309
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2070320142
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40