bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0764_orf3
Length=153
Score E
Sequences producing significant alignments: (Bits) Value
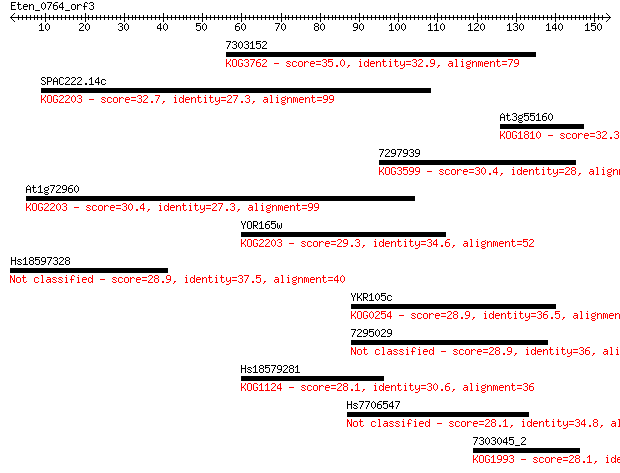
7303152 35.0 0.049
SPAC222.14c 32.7 0.26
At3g55160 32.3 0.33
7297939 30.4 1.4
At1g72960 30.4 1.5
YOR165w 29.3 2.7
Hs18597328 28.9 3.5
YKR105c 28.9 3.7
7295029 28.9 4.4
Hs18579281 28.1 6.5
Hs7706547 28.1 7.0
7303045_2 28.1 7.0
> 7303152
Length=762
Score = 35.0 bits (79), Expect = 0.049, Method: Composition-based stats.
Identities = 26/84 (30%), Positives = 44/84 (52%), Gaps = 7/84 (8%)
Query 56 SWRSVPLWGWLLLICLGWNEIGMV-----VSFATSNWLILPLLLLGLLLGVAVLVSGQMG 110
S+R + G + ++CLG IG V +S+ T+ W++LP L+ + AV +
Sbjct 561 SFRLITQIGHVKVLCLGL--IGNVLRFLYISYLTNPWMVLPFELMQGITHAAVWAASCSY 618
Query 111 IATHSAKHLTQLVRLLLQPFLYGL 134
IA ++ KHL + +LQ +GL
Sbjct 619 IAHNTPKHLRASAQGVLQGIHHGL 642
> SPAC222.14c
Length=762
Score = 32.7 bits (73), Expect = 0.26, Method: Composition-based stats.
Identities = 27/102 (26%), Positives = 44/102 (43%), Gaps = 16/102 (15%)
Query 9 EQPMPAAFYRPLIDSISEQAIQRRAVASMQQQC-REAQLLQQRCGSGA--SWRSVPLWGW 65
E PA+F+ L RR V+ + R A L+ C + +P + W
Sbjct 613 EYTSPASFFTIL---------NRRRVSDISVNFKRSADLIFMDCKRSVINTTTRIPPYFW 663
Query 66 LLLICLGWNEIGMVVSFATSNWLILPLLLLGLLLGVAVLVSG 107
+LLI LGWNE ++ N + +L+ G + + +SG
Sbjct 664 VLLIVLGWNEFMAIL----RNPFVFMILMFGGTVVYGLYISG 701
> At3g55160
Length=2149
Score = 32.3 bits (72), Expect = 0.33, Method: Composition-based stats.
Identities = 14/21 (66%), Positives = 16/21 (76%), Gaps = 0/21 (0%)
Query 126 LLQPFLYGLLKRATDLMDPSG 146
LL PF+Y LK ATDL+D SG
Sbjct 1323 LLHPFIYSELKAATDLLDTSG 1343
> 7297939
Length=897
Score = 30.4 bits (67), Expect = 1.4, Method: Composition-based stats.
Identities = 14/59 (23%), Positives = 28/59 (47%), Gaps = 9/59 (15%)
Query 95 LGLLLGVAVLVSGQMGIATHSAKH---------LTQLVRLLLQPFLYGLLKRATDLMDP 144
L+ G+ L Q+G+ KH + ++R++L F Y L+++A ++ P
Sbjct 689 FSLMFGIVFLAYAQLGLLLFGTKHPDFRNFITSILTMIRMILGDFQYNLIEQANRVLGP 747
> At1g72960
Length=748
Score = 30.4 bits (67), Expect = 1.5, Method: Composition-based stats.
Identities = 27/99 (27%), Positives = 46/99 (46%), Gaps = 7/99 (7%)
Query 5 ATSEEQPMPAAFYRPLIDSISEQAIQRRAVASMQQQCREAQLLQQRCGSGASWRSVPLWG 64
A+S +P++ R LI + ++I R+ + +A Q+ G +W P W
Sbjct 584 ASSTWDEVPSS--RTLITPVQCKSIWRQFKTETEYTVTQAISAQEANRRGNNWLPPP-WA 640
Query 65 WLLLICLGWNEIGMVVSFATSNWLILPLLLLGLLLGVAV 103
L LI LG+NE ++ N L L ++ + LL A+
Sbjct 641 ILALIVLGFNEFMTLL----RNPLYLGVMFVAFLLAKAL 675
> YOR165w
Length=776
Score = 29.3 bits (64), Expect = 2.7, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 26/53 (49%), Gaps = 9/53 (16%)
Query 60 VPLWGWLLLICLGWNEIGMVVSFATSNWLILPLLL-LGLLLGVAVLVSGQMGI 111
+P W ++LL LGWNE V+ PL + L L+LG V + G+
Sbjct 682 IPPWIYVLLAVLGWNEFVAVIRN--------PLFVTLTLILGATFFVIHKFGL 726
> Hs18597328
Length=647
Score = 28.9 bits (63), Expect = 3.5, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query 1 AQAHATSEEQPMPAAFYRPLIDSISEQAIQRRAVASMQQQ 40
A+ H QP+PA Y PL I E++ RR A +Q++
Sbjct 210 AECHRQFRAQPVPAHVYLPLYQEIMERSEARRQ-AGIQKR 248
> YKR105c
Length=582
Score = 28.9 bits (63), Expect = 3.7, Method: Composition-based stats.
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 2/52 (3%)
Query 88 LILPLLLLGLLLGVAVLVSGQMGIATHSAKHLTQLVRLLLQPFLYGLLKRAT 139
L+ PLL+ G +LGV + +G M + T+++ TQ+ LLL F G +A+
Sbjct 399 LVKPLLIFGGVLGV--IGAGLMTLMTNTSTKSTQIGVLLLPGFSLGFALQAS 448
> 7295029
Length=1274
Score = 28.9 bits (63), Expect = 4.4, Method: Composition-based stats.
Identities = 18/50 (36%), Positives = 30/50 (60%), Gaps = 7/50 (14%)
Query 88 LILPLLLLGLLLGVAVLVSGQMGIATHSAKHLTQLVRLLLQPFLYGLLKR 137
+++P+LL+ LL+G+AV G + S + QL RL +Q L+ L+R
Sbjct 1069 ILMPILLMNLLIGLAV---GDI----ESVRRNAQLKRLAMQVVLHTELER 1111
> Hs18579281
Length=733
Score = 28.1 bits (61), Expect = 6.5, Method: Composition-based stats.
Identities = 11/36 (30%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 60 VPLWGWLLLICLGWNEIGMVVSFATSNWLILPLLLL 95
VP G+ +L+ GW +I F +W+ L +++L
Sbjct 199 VPSMGFCILVAHGWQKISTKSVFKKLSWICLSMVIL 234
> Hs7706547
Length=704
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 16/46 (34%), Positives = 26/46 (56%), Gaps = 0/46 (0%)
Query 87 WLILPLLLLGLLLGVAVLVSGQMGIATHSAKHLTQLVRLLLQPFLY 132
+L+ P L++ LL VAVLV G+ T S + + L ++ + F Y
Sbjct 224 YLLQPQLIVRRLLDVAVLVPGRPSEQTLSPHNRSALYKVFVPSFTY 269
> 7303045_2
Length=1053
Score = 28.1 bits (61), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 6/33 (18%)
Query 119 LTQLVRLL------LQPFLYGLLKRATDLMDPS 145
L QLVR + ++PFLY +++ +TDL PS
Sbjct 655 LEQLVRTIRDVPEPMKPFLYSVIELSTDLQQPS 687
Lambda K H
0.324 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1961355924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40