bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0741_orf1
Length=124
Score E
Sequences producing significant alignments: (Bits) Value
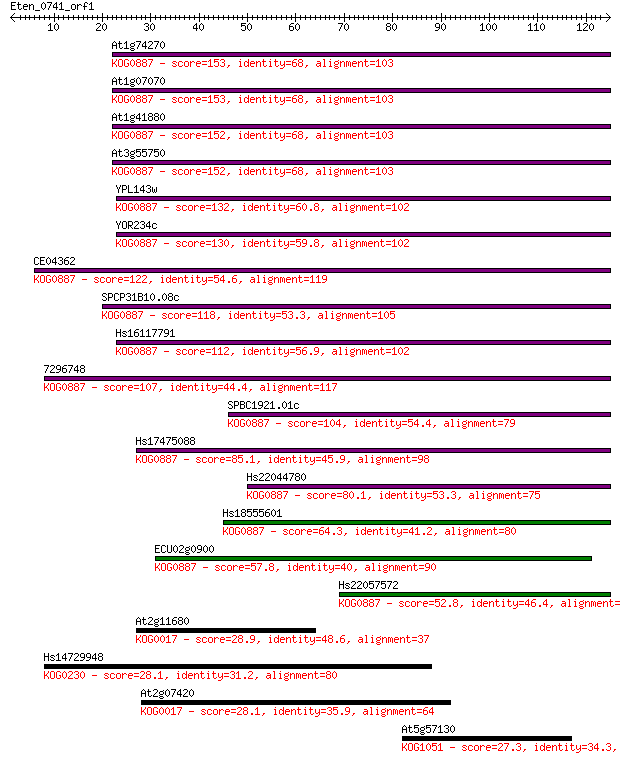
At1g74270 153 7e-38
At1g07070 153 7e-38
At1g41880 152 1e-37
At3g55750 152 1e-37
YPL143w 132 2e-31
YOR234c 130 6e-31
CE04362 122 2e-28
SPCP31B10.08c 118 3e-27
Hs16117791 112 1e-25
7296748 107 7e-24
SPBC1921.01c 104 3e-23
Hs17475088 85.1 3e-17
Hs22044780 80.1 1e-15
Hs18555601 64.3 6e-11
ECU02g0900 57.8 5e-09
Hs22057572 52.8 2e-07
At2g11680 28.9 2.2
Hs14729948 28.1 3.6
At2g07420 28.1 4.1
At5g57130 27.3 7.5
> At1g74270
Length=112
Score = 153 bits (387), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 70/103 (67%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 22 VRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKF 81
VRLYVRG +LGYKRSK NQ P+ SL+Q+E V T+++ +Y GKR+AY+YKAKT+K G+ +
Sbjct 10 VRLYVRGTILGYKRSKSNQYPNTSLVQIEGVNTQEEVNWYKGKRMAYIYKAKTKKNGSHY 69
Query 82 RTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
R IWGK+ RPHGNSGVVRA+F NLPPKS G RVRVF+YPSNI
Sbjct 70 RCIWGKVTRPHGNSGVVRAKFTSNLPPKSMGSRVRVFMYPSNI 112
> At1g07070
Length=112
Score = 153 bits (386), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 70/103 (67%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 22 VRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKF 81
VRLYVRG +LGYKRSK NQ P+ SL+Q+E V T ++ ++Y GKR+AY+YKAKT+K G+ +
Sbjct 10 VRLYVRGTILGYKRSKSNQYPNTSLVQVEGVNTTEEVSWYKGKRMAYIYKAKTKKNGSHY 69
Query 82 RTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
R IWGK+ RPHGNSGVVRA+F NLPPKS G RVRVF+YPSNI
Sbjct 70 RCIWGKVTRPHGNSGVVRAKFTSNLPPKSMGSRVRVFMYPSNI 112
> At1g41880
Length=111
Score = 152 bits (385), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 70/103 (67%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 22 VRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKF 81
VRLYVRG VLGYKRSK NQ P+ SL+Q+E V T+++ +Y GKR+AY+YKAKT+K G+ +
Sbjct 9 VRLYVRGTVLGYKRSKSNQYPNTSLIQIEGVNTQEEVNWYKGKRLAYIYKAKTKKNGSHY 68
Query 82 RTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
R IWGK+ RPHGNSGVVR++F NLPPKS G RVRVF+YPSNI
Sbjct 69 RCIWGKVTRPHGNSGVVRSKFTSNLPPKSMGARVRVFMYPSNI 111
> At3g55750
Length=111
Score = 152 bits (385), Expect = 1e-37, Method: Compositional matrix adjust.
Identities = 70/103 (67%), Positives = 86/103 (83%), Gaps = 0/103 (0%)
Query 22 VRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKF 81
VRLYVRG VLGYKRSK NQ P+ SL+Q+E V T+++ +Y GKR+AY+YKAKT+K G+ +
Sbjct 9 VRLYVRGTVLGYKRSKSNQYPNTSLVQIEGVNTQEEVNWYKGKRLAYIYKAKTKKNGSHY 68
Query 82 RTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
R IWGK+ RPHGNSGVVR++F NLPPKS G RVRVF+YPSNI
Sbjct 69 RCIWGKVTRPHGNSGVVRSKFTSNLPPKSMGARVRVFMYPSNI 111
> YPL143w
Length=107
Score = 132 bits (332), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 62/102 (60%), Positives = 77/102 (75%), Gaps = 0/102 (0%)
Query 23 RLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKFR 82
RLYV+G L Y+RSK P+ SL+++E V T +D FYLGKR+AYVY+A E +G+K R
Sbjct 6 RLYVKGKHLSYQRSKRVNNPNVSLIKIEGVATPQDAQFYLGKRIAYVYRASKEVRGSKIR 65
Query 83 TIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
+WGK+ R HGNSGVVRA FR NLP K+FG VR+FLYPSNI
Sbjct 66 VMWGKVTRTHGNSGVVRATFRNNLPAKTFGASVRIFLYPSNI 107
> YOR234c
Length=107
Score = 130 bits (327), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 61/102 (59%), Positives = 77/102 (75%), Gaps = 0/102 (0%)
Query 23 RLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKFR 82
RLYV+G L Y+RSK P+ SL+++E V T ++ FYLGKR+AYVY+A E +G+K R
Sbjct 6 RLYVKGKHLSYQRSKRVNNPNVSLIKIEGVATPQEAQFYLGKRIAYVYRASKEVRGSKIR 65
Query 83 TIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
+WGK+ R HGNSGVVRA FR NLP K+FG VR+FLYPSNI
Sbjct 66 VMWGKVTRTHGNSGVVRATFRNNLPAKTFGASVRIFLYPSNI 107
> CE04362
Length=124
Score = 122 bits (305), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 65/125 (52%), Positives = 81/125 (64%), Gaps = 8/125 (6%)
Query 6 AEASQKRRPTKGTKQPVRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKR 65
A+ + RRP+ T RLYV+ I G+KR Q+ SLL+LE V ++D FY GKR
Sbjct 2 ADTAVARRPSAPTTG--RLYVKAIFTGFKRGLRTQSEHTSLLKLEGVFNKEDAGFYAGKR 59
Query 66 VAYVYKA--KTEKQG----TKFRTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFL 119
V Y+YKA KT K G T+ R IWGKI RPHGN+G VRA+F N+PP + G R+RV L
Sbjct 60 VVYLYKAHNKTLKTGHTVATRTRAIWGKITRPHGNAGAVRAKFHHNIPPSALGKRIRVLL 119
Query 120 YPSNI 124
YPSNI
Sbjct 120 YPSNI 124
> SPCP31B10.08c
Length=108
Score = 118 bits (295), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 56/105 (53%), Positives = 72/105 (68%), Gaps = 0/105 (0%)
Query 20 QPVRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGT 79
Q RLYV+ L ++RSK P S++++E ++++ FYLGKRV YVYK+ +G+
Sbjct 4 QGHRLYVKAKHLSFQRSKHVIHPGTSIVKIEGCDSKEEAQFYLGKRVCYVYKSSKAVRGS 63
Query 80 KFRTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
K R IWG I RPHGNSG VRARF NLP K+FG +RV LYPSNI
Sbjct 64 KIRVIWGTIARPHGNSGAVRARFVHNLPAKTFGSSLRVMLYPSNI 108
> Hs16117791
Length=110
Score = 112 bits (281), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 58/107 (54%), Positives = 70/107 (65%), Gaps = 5/107 (4%)
Query 23 RLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEK-----Q 77
RL+ + I GYKR NQ +LL++E V R +T FYLGKR AYVYKAK +
Sbjct 4 RLWSKAIFAGYKRGLRNQREHTALLKIEGVYARDETEFYLGKRCAYVYKAKNNTVTPGGK 63
Query 78 GTKFRTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
K R IWGK+ R HGNSG+VRA+FR NLP K+ G R+RV LYPS I
Sbjct 64 PNKTRVIWGKVTRAHGNSGMVRAKFRSNLPAKAIGHRIRVMLYPSRI 110
> 7296748
Length=157
Score = 107 bits (266), Expect = 7e-24, Method: Compositional matrix adjust.
Identities = 52/124 (41%), Positives = 80/124 (64%), Gaps = 7/124 (5%)
Query 8 ASQKRRPTKGTKQPVRLYVRGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVA 67
AS+ + K K+ RL+ + + GYKR NQ ++++L++E + ++ +FY+GKR
Sbjct 34 ASEAKVSAKKYKRHGRLFAKAVFTGYKRGLRNQHENQAILKIEGARRKEHGSFYVGKRCV 93
Query 68 YVYKAKTEK-------QGTKFRTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLY 120
YVYKA+T+K + T+ R +WGK+ R HGN+G VRARF +NLP + G R+R+ LY
Sbjct 94 YVYKAETKKCVPQHPERKTRVRAVWGKVTRIHGNTGAVRARFNRNLPGHAMGHRIRIMLY 153
Query 121 PSNI 124
PS I
Sbjct 154 PSRI 157
> SPBC1921.01c
Length=79
Score = 104 bits (260), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 43/79 (54%), Positives = 60/79 (75%), Gaps = 0/79 (0%)
Query 46 LLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKFRTIWGKICRPHGNSGVVRARFRKN 105
++++E ++++ FYLGKR+ +VYK+ +G+K R IWG + RPHGNSGVVRARF N
Sbjct 1 MVKIEGCDSKEEAQFYLGKRICFVYKSNKPVRGSKIRVIWGTVSRPHGNSGVVRARFTHN 60
Query 106 LPPKSFGGRVRVFLYPSNI 124
LPPK+FG +RV LYPSN+
Sbjct 61 LPPKTFGASLRVMLYPSNV 79
> Hs17475088
Length=138
Score = 85.1 bits (209), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 45/103 (43%), Positives = 63/103 (61%), Gaps = 5/103 (4%)
Query 27 RGIVLGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEK-----QGTKF 81
+ I GYK+ NQ +LL++E+V + +T F LGKR AY+YKAK + +
Sbjct 36 KAIFAGYKQGLWNQREHTALLKIEDVYAQDETEFCLGKRSAYLYKAKNNTVTPGGKPNQT 95
Query 82 RTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
R IWGK+ + +GNSGVV A+F+ LP K+ + V LYPSNI
Sbjct 96 RVIWGKVTQTYGNSGVVCAKFQSKLPAKATRHGIHVMLYPSNI 138
> Hs22044780
Length=143
Score = 80.1 bits (196), Expect = 1e-15, Method: Compositional matrix adjust.
Identities = 40/80 (50%), Positives = 50/80 (62%), Gaps = 5/80 (6%)
Query 50 ENVKTRKDTAFYLGKRVAYVYKAKT-----EKQGTKFRTIWGKICRPHGNSGVVRARFRK 104
E V +R + FYL KR YVYKAK + + K R IWGK+ HGNSG+V A+F+
Sbjct 64 EGVYSRDEAEFYLSKRCVYVYKAKNNTVTPDGKPNKTRVIWGKVTLAHGNSGMVHAKFQS 123
Query 105 NLPPKSFGGRVRVFLYPSNI 124
NL K+ G RVRV L+PS I
Sbjct 124 NLTAKAIGHRVRVMLHPSRI 143
> Hs18555601
Length=108
Score = 64.3 bits (155), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 33/83 (39%), Positives = 47/83 (56%), Gaps = 3/83 (3%)
Query 45 SLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTK---FRTIWGKICRPHGNSGVVRAR 101
++L++E V R +T FYLG R YV K + G+K R IWGK+ G + VV A+
Sbjct 23 TVLEIEGVYARDETEFYLGNRCTYVCKEQHSGPGSKSNKTRVIWGKVTCAQGKNSVVCAK 82
Query 102 FRKNLPPKSFGGRVRVFLYPSNI 124
F+ + P K+ G R+ V LY I
Sbjct 83 FQSHPPAKAIGHRIHVMLYHWRI 105
> ECU02g0900
Length=113
Score = 57.8 bits (138), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/90 (40%), Positives = 50/90 (55%), Gaps = 4/90 (4%)
Query 31 LGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKFRTIWGKICR 90
+ +KR + P LL++ NV ++ + + Y+G V YK E G ++ + G I R
Sbjct 21 VSFKRGQRRLHPRHVLLKIRNVTSKDEASSYVGNGVV-CYKKNNE--GERY-PVHGVITR 76
Query 91 PHGNSGVVRARFRKNLPPKSFGGRVRVFLY 120
HGNSG VRARF +NL PK G V V LY
Sbjct 77 IHGNSGAVRARFTRNLDPKLLGTHVFVKLY 106
> Hs22057572
Length=205
Score = 52.8 bits (125), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 26/59 (44%), Positives = 36/59 (61%), Gaps = 3/59 (5%)
Query 69 VYKAKTEKQG---TKFRTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVRVFLYPSNI 124
V+K T G K IWGK+ + HGN G+V ++F+ N P K+ G ++RV LYPS I
Sbjct 147 VWKNNTVTPGGKPNKAIIIWGKVIQAHGNGGMVCSKFQSNPPAKAIGHKIRVMLYPSRI 205
> At2g11680
Length=301
Score = 28.9 bits (63), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 18/41 (43%), Positives = 24/41 (58%), Gaps = 4/41 (9%)
Query 27 RGIVLGYKRSK----VNQTPSRSLLQLENVKTRKDTAFYLG 63
GIVLG+K SK VN+ ++QL+ KT KD +LG
Sbjct 4 EGIVLGHKISKKGVQVNKAKVDVMMQLQPPKTGKDIRSFLG 44
> Hs14729948
Length=558
Score = 28.1 bits (61), Expect = 3.6, Method: Composition-based stats.
Identities = 25/81 (30%), Positives = 39/81 (48%), Gaps = 15/81 (18%)
Query 8 ASQKRRPTKGTKQPVRLYVRGIV-LGYKRSKVNQTPSRSLLQLENVKTRKDTAFYLGKRV 66
A Q++RPT K + G+ +GYK S+ N LL +EN+ FY G+++
Sbjct 361 AVQQKRPTALAK------ILGVYRIGYKNSQNNTEKKLDLLVMENL-------FY-GRKM 406
Query 67 AYVYKAKTEKQGTKFRTIWGK 87
A V+ K + +T GK
Sbjct 407 AQVFDLKGSLRNRNVKTDTGK 427
> At2g07420
Length=1664
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 23/68 (33%), Positives = 33/68 (48%), Gaps = 14/68 (20%)
Query 28 GIVLGYKRSK----VNQTPSRSLLQLENVKTRKDTAFYLGKRVAYVYKAKTEKQGTKFRT 83
GIVLG+K SK V++ + ++QL+ KT KD +LG Y +F
Sbjct 1523 GIVLGHKISKKGIEVDKGKIKVIMQLQPPKTVKDIRSFLGHAGFY----------RRFIK 1572
Query 84 IWGKICRP 91
+ KI RP
Sbjct 1573 DFSKIARP 1580
> At5g57130
Length=1028
Score = 27.3 bits (59), Expect = 7.5, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 18/35 (51%), Gaps = 0/35 (0%)
Query 82 RTIWGKICRPHGNSGVVRARFRKNLPPKSFGGRVR 116
R + + C P G++ A FR+ P + GG VR
Sbjct 914 RFVLNRSCEPGIEKGMITAAFREIFPEREEGGGVR 948
Lambda K H
0.320 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1187579072
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40