bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0734_orf1
Length=199
Score E
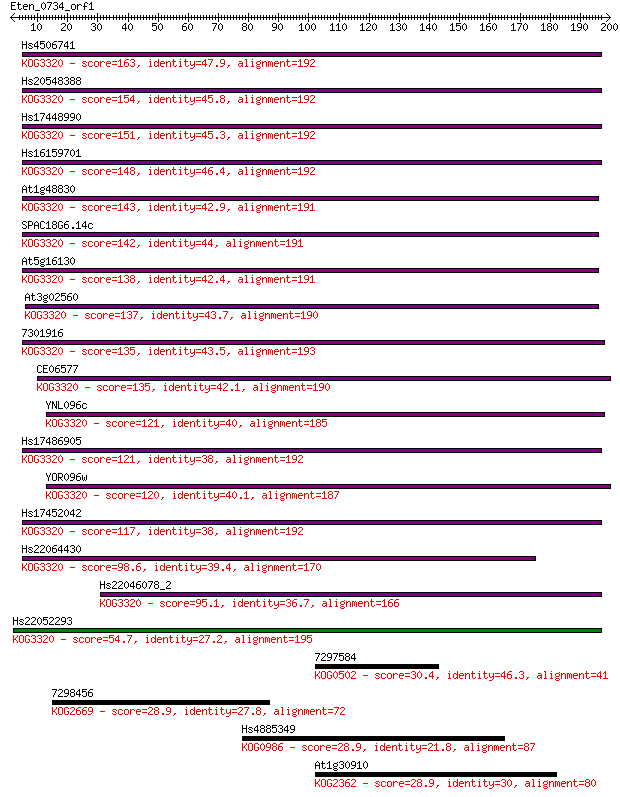
Sequences producing significant alignments: (Bits) Value
Hs4506741 163 2e-40
Hs20548388 154 9e-38
Hs17448990 151 9e-37
Hs16159701 148 9e-36
At1g48830 143 2e-34
SPAC18G6.14c 142 4e-34
At5g16130 138 8e-33
At3g02560 137 1e-32
7301916 135 5e-32
CE06577 135 6e-32
YNL096c 121 8e-28
Hs17486905 121 8e-28
YOR096w 120 1e-27
Hs17452042 117 1e-26
Hs22064430 98.6 7e-21
Hs22046078_2 95.1 8e-20
Hs22052293 54.7 1e-07
7297584 30.4 2.7
7298456 28.9 6.4
Hs4885349 28.9 6.9
At1g30910 28.9 7.3
> Hs4506741
Length=194
Score = 163 bits (412), Expect = 2e-40, Method: Compositional matrix adjust.
Identities = 92/196 (46%), Positives = 132/196 (67%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV G+
Sbjct 2 FSSSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEV-GGGRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLT V DA+LED+V PSE+VG+R RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSRTLTAVHDAILEDLVFPSEIVGKRIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVNFEFPE 191
> Hs20548388
Length=194
Score = 154 bits (390), Expect = 9e-38, Method: Compositional matrix adjust.
Identities = 88/196 (44%), Positives = 129/196 (65%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +IVK + +P E E + + L ++E + S+L+A++ EL I A K +E+ G+
Sbjct 2 FSSSAKIVKPNDEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEL-GGGRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++IFVP + ++I RLV+ELEKK +HVV I QR I+ ++ KN + RP
Sbjct 59 AIIIFVPI-PQLKSFQKIQVRLVRELEKKFSGKHVVFIVQRRILPKPTRKSRTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RS TLT V DA+LED+V PSE+VG+R RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSHTLTAVHDAILEDLVFPSEIVGKRIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+LT KD F FPE
Sbjct 176 VYKKLTGKDVNFEFPE 191
> Hs17448990
Length=194
Score = 151 bits (381), Expect = 9e-37, Method: Compositional matrix adjust.
Identities = 87/196 (44%), Positives = 127/196 (64%), Gaps = 10/196 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +IVK +G +P E E + + L ++E + SEL+A++ EL I A K +EV G+
Sbjct 2 FSSSAKIVKPNGEKPEEFESGISQALLELEMN--SELKAQLRELNITAAKEIEV-GGGRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKR---KNLKVRP 120
A++I VP + ++I RLV ELEKK +HVV IAQR I+ ++ KN + RP
Sbjct 59 AIIILVPV-TQLKSFQKIQIRLVSELEKKFSGKHVVFIAQRRILPKPTRKSCTKNKQKRP 117
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RS T+T V DA+LED+V PSE+VG++ RV+ DGS+L+KVHLD ++N+E K+ TF+
Sbjct 118 RSSTVTAVHDAILEDLVFPSEIVGKKIRVKLDGSRLIKVHLD--KAQQNNVEHKVETFSG 175
Query 181 VYKRLTNKDAAFLFPE 196
VYK+L KD FPE
Sbjct 176 VYKKLMGKDVNCEFPE 191
> Hs16159701
Length=194
Score = 148 bits (373), Expect = 9e-36, Method: Compositional matrix adjust.
Identities = 89/200 (44%), Positives = 126/200 (63%), Gaps = 18/200 (9%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+T +IVK +G +P E E + + L ++E + +L+A++ EL I A K +EV G+
Sbjct 2 FSTSAKIVKPNGEKPDEFESRISQALLELEMNL--DLKAQLRELNIMAAKEIEV-GGGQK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIV-------GLDYKRKNL 116
A++IFVP + ++I RLV ELEKK +HVV IAQR I+ YK+K+
Sbjct 59 AIIIFVPVP-QLKSFQKIQVRLVCELEKKFSGKHVVFIAQRRILPKPTWKSCTKYKQKH- 116
Query 117 KVRPRSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLW 176
PRS TLT V DA+LED+V S++VG+R RV+ DGS+LMKVHLD ++N+E K+
Sbjct 117 ---PRSHTLTAVHDAILEDLVFTSKIVGKRIRVKLDGSRLMKVHLD--KAQQNNVEHKVE 171
Query 177 TFAAVYKRLTNKDAAFLFPE 196
TF+ VYK+L KD F FPE
Sbjct 172 TFSGVYKKLMGKDVNFEFPE 191
> At1g48830
Length=191
Score = 143 bits (361), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 82/192 (42%), Positives = 131/192 (68%), Gaps = 7/192 (3%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+ + +I K G + +E++++V + D+E++ Q EL++E+ +L + + V++ + G+
Sbjct 2 FSAQHKIHKEKGVELSELDEQVAQAFFDLENTNQ-ELKSELKDLYVNSAVQVDI-SGGRK 59
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+V+ VPYR+ ++IH RLV+ELEKK + V++IA R IV K K RPR+R
Sbjct 60 AIVVNVPYRL-RKAYRKIHVRLVRELEKKFSGKDVILIATRRIVRPPKKGSAAK-RPRNR 117
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT+V +A+L+D+V P+E+VG+R R R DG+K+MKV LDPK+ ++N E K+ F+AVYK
Sbjct 118 TLTSVHEAILDDVVLPAEIVGKRTRYRLDGTKIMKVFLDPKE--RNNTEYKVEAFSAVYK 175
Query 184 RLTNKDAAFLFP 195
+LT KD F FP
Sbjct 176 KLTGKDVVFEFP 187
> SPAC18G6.14c
Length=195
Score = 142 bits (359), Expect = 4e-34, Method: Compositional matrix adjust.
Identities = 84/193 (43%), Positives = 124/193 (64%), Gaps = 7/193 (3%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
M+ +IVK SQPTE + V +CL D+ESS++ ++ E+ L I + + VEV GK
Sbjct 1 MSALNKIVKRSSSQPTETDLLVAQCLYDLESSSK-DMAKELRPLQITSAREVEV-GGGKK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNL-KVRPRS 122
A+V+FVP + + + ARL +ELEKK RHV+ IAQR I+ ++ + + RPRS
Sbjct 59 AIVVFVP-QPLLKAFHKCQARLTRELEKKFADRHVIFIAQRRILPKPGRKSRVTQKRPRS 117
Query 123 RTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVY 182
RTLT V +A+LEDIV P+E++G+R R DG K +KV LD +D + ++ KL +F++VY
Sbjct 118 RTLTAVHNAILEDIVFPTEIIGKRTRQATDGRKTIKVFLDNRD--ANTVDYKLGSFSSVY 175
Query 183 KRLTNKDAAFLFP 195
+LT K+ F FP
Sbjct 176 HKLTGKNVTFEFP 188
> At5g16130
Length=190
Score = 138 bits (347), Expect = 8e-33, Method: Compositional matrix adjust.
Identities = 81/192 (42%), Positives = 126/192 (65%), Gaps = 7/192 (3%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
+ + +I K ++PTE E++V + L D+E++ Q EL++E+ +L I +++ + K
Sbjct 2 FSAQNKIKKDKNAEPTECEEQVAQALFDLENTNQ-ELKSELKDLYINQAVHMDISGNRK- 59
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+VI+VP+R+ ++IH RLV+ELEKK + V+ + R I+ K ++ RPR+R
Sbjct 60 AVVIYVPFRL-RKAFRKIHPRLVRELEKKFSGKDVIFVTTRRIMRPPKKGAAVQ-RPRNR 117
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT+V +AMLED+ P+E+VG+R R R DGSK+MKV LD K+ K+N E KL T VY+
Sbjct 118 TLTSVHEAMLEDVAFPAEIVGKRTRYRLDGSKIMKVFLDAKE--KNNTEYKLETMVGVYR 175
Query 184 RLTNKDAAFLFP 195
+LT KD F +P
Sbjct 176 KLTGKDVVFEYP 187
> At3g02560
Length=191
Score = 137 bits (345), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 83/192 (43%), Positives = 127/192 (66%), Gaps = 9/192 (4%)
Query 6 TTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASG-KT 64
+ + +I K G PTE E++V + L D+E++ Q EL++E+ +L I + V++ SG +
Sbjct 3 SGQNKIHKDKGVAPTEFEEQVTQALFDLENTNQ-ELKSELKDLYIN--QAVQMDISGNRK 59
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A+VI+VP+R+ ++IH RLV+ELEKK + V+ +A R I+ K ++ RPR+R
Sbjct 60 AVVIYVPFRL-RKAFRKIHLRLVRELEKKFSGKDVIFVATRRIMRPPKKGSAVQ-RPRNR 117
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYK 183
TLT+V +AMLED+ P+E+VG+R R R DG+K+MKV LD K K++ E KL T VY+
Sbjct 118 TLTSVHEAMLEDVAYPAEIVGKRTRYRLDGTKIMKVFLD--SKLKNDTEYKLETMVGVYR 175
Query 184 RLTNKDAAFLFP 195
+LT KD F +P
Sbjct 176 KLTGKDVVFEYP 187
> 7301916
Length=194
Score = 135 bits (341), Expect = 5e-32, Method: Compositional matrix adjust.
Identities = 84/197 (42%), Positives = 124/197 (62%), Gaps = 11/197 (5%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
M +I+K GS P + EK + + L ++E A S+L+ + +L I + +E K
Sbjct 1 MAIGSKIIKPGGSDPDDFEKSIAQALVELE--ANSDLKPYLRDLHITRAREIEF--GSKK 56
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK--RKNLKV-RP 120
A++I+VP V ++I LV+ELEKK +HVV+IA+R I+ + R LK RP
Sbjct 57 AVIIYVPIP-QQKVFQKIQIILVRELEKKFSGKHVVVIAERKILPKPTRKARNPLKQKRP 115
Query 121 RSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAA 180
RSRTLT V DA+LED+V P+E+VG+R RV+ DGS+L+KVHLD + +E K+ TF +
Sbjct 116 RSRTLTAVYDAILEDLVFPAEIVGKRIRVKLDGSQLVKVHLDKNQQT--TIEHKVDTFTS 173
Query 181 VYKRLTNKDAAFLFPEH 197
VYK+LT +D F FP++
Sbjct 174 VYKKLTGRDVTFEFPDN 190
> CE06577
Length=194
Score = 135 bits (340), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 80/195 (41%), Positives = 126/195 (64%), Gaps = 12/195 (6%)
Query 10 RIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIF 69
+++K DG +E+EK+V + L D+E++ ++++++ EL I VK VE+ K+A++I+
Sbjct 7 KLLKSDGKVVSEIEKQVSQALIDLETN--DDVQSQLKELYIVGVKEVEL--GNKSAIIIY 62
Query 70 VPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK----RKNLKVRPRSRT 124
VP + +IH LV+ELEKK R ++I+A+R I+ + R + RPRSRT
Sbjct 63 VPVP-QLKAFHKIHPALVRELEKKFGGRDILILAKRRILPKPQRGSKARPQKQKRPRSRT 121
Query 125 LTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKR 184
LT V DA L+++V P+EVVGRR RV+ DG K+ KVHLD + N+ K+ FA+VY++
Sbjct 122 LTAVHDAWLDELVYPAEVVGRRIRVKLDGKKVYKVHLDKSHQT--NVGHKIGVFASVYRK 179
Query 185 LTNKDAAFLFPEHVY 199
LT KD F FP+ ++
Sbjct 180 LTGKDVTFEFPDPIF 194
> YNL096c
Length=190
Score = 121 bits (304), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 74/187 (39%), Positives = 116/187 (62%), Gaps = 7/187 (3%)
Query 13 KVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPY 72
K+ P+E+E +V + D+ESS+ EL+A++ L I +++ ++V GK ALV+FVP
Sbjct 7 KILSQAPSELELQVAKTFIDLESSS-PELKADLRPLQIKSIREIDV-TGGKKALVLFVPV 64
Query 73 RVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK-RKNLKVRPRSRTLTNVQD 130
++ ++ +L +ELEKK RHV+ +A+R I+ + + ++ RPRSRTLT V D
Sbjct 65 PA-LSAYHKVQTKLTRELEKKFPDRHVIFLAERRILPKPSRTSRQVQKRPRSRTLTAVHD 123
Query 131 AMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDA 190
+LED+V P+E+VG+R R G+K+ KV LD KD + ++ KL +F AVY +LT K
Sbjct 124 KVLEDMVFPTEIVGKRVRYLVGGNKIQKVLLDSKDVQQ--IDYKLESFQAVYNKLTGKQI 181
Query 191 AFLFPEH 197
F P
Sbjct 182 VFEIPSQ 188
> Hs17486905
Length=176
Score = 121 bits (304), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 73/192 (38%), Positives = 113/192 (58%), Gaps = 20/192 (10%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++R +IVK +G +P E E + + L ++E + +L+A++ EL I A K +EV G+
Sbjct 2 FSSRAKIVKPNGEKPDEFESGISQALLELEMNL--DLKAQLWELNITAAKEIEV-GGGRK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRT 124
A++IFVP + Q+ + +L+R + Q+S N + PRS T
Sbjct 59 AIIIFVPVP---------QLKSFQKTQVQLRRILPKPTQKSCTN------NKQKLPRSCT 103
Query 125 LTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKR 184
LT V DA+LED+V PSE+VG+R V+ DGS L+K+HLD + ++N+E K+ F+ VYK+
Sbjct 104 LTAVHDAILEDLVFPSEIVGKRIHVKLDGSHLIKIHLD--EAQQNNVEHKVEPFSGVYKK 161
Query 185 LTNKDAAFLFPE 196
L KD F FPE
Sbjct 162 LMGKDVNFEFPE 173
> YOR096w
Length=190
Score = 120 bits (302), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 75/189 (39%), Positives = 114/189 (60%), Gaps = 7/189 (3%)
Query 13 KVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPY 72
K+ PTE+E +V + ++E+S+ EL+AE+ L +++ ++V A GK AL IFVP
Sbjct 7 KILSQAPTELELQVAQAFVELENSS-PELKAELRPLQFKSIREIDV-AGGKKALAIFVPV 64
Query 73 RVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYK-RKNLKVRPRSRTLTNVQD 130
K + +L +ELEKK + RHV+ +A+R I+ + + ++ RPRSRTLT V D
Sbjct 65 PSLAGFHK-VQTKLTRELEKKFQDRHVIFLAERRILPKPSRTSRQVQKRPRSRTLTAVHD 123
Query 131 AMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDA 190
+LED+V P+E+VG+R R G+K+ KV LD KD ++ KL +F AVY +LT K
Sbjct 124 KILEDLVFPTEIVGKRVRYLVGGNKIQKVLLDSKD--VQQIDYKLESFQAVYNKLTGKQI 181
Query 191 AFLFPEHVY 199
F P +
Sbjct 182 VFEIPSETH 190
> Hs17452042
Length=191
Score = 117 bits (293), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 73/192 (38%), Positives = 117/192 (60%), Gaps = 20/192 (10%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +I+K + +P E++ + + L +E S S+L+A++ EL I A K EV SG+
Sbjct 17 FSSSAKIMKPNDEKPDELKPSISQALLQLEMS--SDLKAQLRELNITAAKETEV-GSGRK 73
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRT 124
A++IFVP V ++ + Q+++ +L+R + ++S + KN + PRSR
Sbjct 74 AIIIFVP-------VPKLKS--FQKIQVQLRRILPKPTRKSCM------KNKQKHPRSRA 118
Query 125 LTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKR 184
LT V DA+LED+V PSE+VG+R ++ DG L+KVHLD ++N+E K+ F+ VYK+
Sbjct 119 LTAVHDAILEDLVFPSEIVGKRICMKLDGGLLVKVHLD--KAQQNNVEHKVEIFSGVYKK 176
Query 185 LTNKDAAFLFPE 196
LT KD F FPE
Sbjct 177 LTGKDVNFEFPE 188
> Hs22064430
Length=152
Score = 98.6 bits (244), Expect = 7e-21, Method: Compositional matrix adjust.
Identities = 67/171 (39%), Positives = 101/171 (59%), Gaps = 22/171 (12%)
Query 5 MTTRQRIVKVDGSQPTEVEKEVDRCLADIESSAQSELRAEVAELTICAVKPVEVPASGKT 64
++ +IVK +G +P E E + + L ++E + S+L+A++ EL I A K +EV G+
Sbjct 2 FSSSAKIVKPNGEKPDEFESGISQALLELEMN--SDLKAQLRELNITAAKEIEV-GGGQK 58
Query 65 ALVIFVPYRVYMNVVKRIHARLVQELEKKLK-RHVVIIAQRSIVGLDYKRKNLKVRPRSR 123
A++IFVP + ++I RLV+ELEKK +HVV IAQR I+ K +SR
Sbjct 59 AIIIFVPV-PQLKSFQKIQVRLVRELEKKFSGKHVVFIAQRRILP--------KPTRKSR 109
Query 124 TLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDNLEEK 174
T ED+V PSE+V +R RV+ DGS+L+K HLD ++N+E K
Sbjct 110 TKN-------EDLVFPSEIVVKRIRVKLDGSRLIKAHLD--KAQQNNVEHK 151
> Hs22046078_2
Length=152
Score = 95.1 bits (235), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 61/166 (36%), Positives = 89/166 (53%), Gaps = 18/166 (10%)
Query 31 ADIESSAQSELRAEVAELTICAVKPVEVPASGKTALVIFVPYRVYMNVVKRIHARLVQEL 90
A +E S L+ + EL I A K E GK A++IFVP+ + ++I L + L
Sbjct 2 ALLELEINSHLKVQFRELNITAAKETEDGGGGK-AIIIFVPF-PQLKSFQKIQVWLRRIL 59
Query 91 EKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRTLTNVQDAMLEDIVAPSEVVGRRWRVR 150
K ++ KN + PRSRTL + D++ ED+V PSE+VG+R V+
Sbjct 60 PKPTRKSCT--------------KNKQKHPRSRTLKAMHDSIPEDLVFPSEIVGKRIHVK 105
Query 151 ADGSKLMKVHLDPKDKAKDNLEEKLWTFAAVYKRLTNKDAAFLFPE 196
DGS+L+K HLD ++N+E + TF+ VY +LT KD FPE
Sbjct 106 LDGSRLIKAHLD--KAQQNNVERNVETFSGVYMKLTGKDINVEFPE 149
> Hs22052293
Length=174
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 53/204 (25%), Positives = 88/204 (43%), Gaps = 46/204 (22%)
Query 2 VQKMTTRQRIVKVDGSQPTEVEKEVDRCLA--DIESSAQSELRAEVAELTICAVKPVEVP 59
++K +T + + + GS P ++ D L+ D ++ +A +A C V +
Sbjct 5 IKKCSTEEVALSLKGSGPARGQEGKDSSLSIRDQMEPCRTRFKAWLATHE-CTVHAPHIQ 63
Query 60 ASG------KTALVIFVPYRVYMNVVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKR 113
+ + ++I VP ++ ++I ++V ELEKK H
Sbjct 64 SRNLKLMVVRKLIIIIVPIP-QLSSFQKIQVQIVHELEKKFATH---------------- 106
Query 114 KNLKVRPRSRTLTNVQDAMLEDIVAPSEVVGRRWRVRADGSKLMKVHLDPKDKAKDN-LE 172
DA+LED+V PSE+ +R V+ +L KVH +DKA+ N +
Sbjct 107 ----------------DAILEDLVCPSEIGSKRICVKPASIQLRKVH---RDKAQQNHVG 147
Query 173 EKLWTFAAVYKRLTNKDAAFLFPE 196
K+ TF+ VYK L K FPE
Sbjct 148 HKVETFSGVYKDLLGKGVNCEFPE 171
> 7297584
Length=237
Score = 30.4 bits (67), Expect = 2.7, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 24/43 (55%), Gaps = 2/43 (4%)
Query 102 AQRSIVGLDYKRKN--LKVRPRSRTLTNVQDAMLEDIVAPSEV 142
A S++ LD KRK+ L RP+S LTN+Q E P EV
Sbjct 26 APTSMLVLDAKRKSAFLPYRPQSTVLTNLQRGNTEATFCPVEV 68
> 7298456
Length=818
Score = 28.9 bits (63), Expect = 6.4, Method: Composition-based stats.
Identities = 20/75 (26%), Positives = 35/75 (46%), Gaps = 3/75 (4%)
Query 15 DGSQPTEVEKEVDRCLADIES---SAQSELRAEVAELTICAVKPVEVPASGKTALVIFVP 71
D S +EKEV+RC+A I + + Q+E+++ A L GK V+
Sbjct 186 DKSHSGNIEKEVERCVAFINAYNKNLQNEIKSRKAVLASIEAAKKFYEHQGKEVKVVASA 245
Query 72 YRVYMNVVKRIHARL 86
Y+ + +K + +L
Sbjct 246 YKSFGTRIKIVKRKL 260
> Hs4885349
Length=590
Score = 28.9 bits (63), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 19/87 (21%), Positives = 37/87 (42%), Gaps = 6/87 (6%)
Query 78 VVKRIHARLVQELEKKLKRHVVIIAQRSIVGLDYKRKNLKVRPRSRTLTNVQDAMLEDIV 137
++K H ++L + + R + + +G R+ + RP D++ E V
Sbjct 33 ILKFPHISQCEDLRRTIDRDYCSLCDKQPIGRLLFRQFCETRPGLECYIQFLDSVAEYEV 92
Query 138 APSEVVGRRWRVRADGSKLMKVHLDPK 164
P E +G + G ++M +L PK
Sbjct 93 TPDEKLGEK------GKEIMTKYLTPK 113
> At1g30910
Length=318
Score = 28.9 bits (63), Expect = 7.3, Method: Compositional matrix adjust.
Identities = 24/90 (26%), Positives = 35/90 (38%), Gaps = 10/90 (11%)
Query 102 AQRSIVGLDYKRKNLKVRPRSRTLTNVQDAMLE--DIVAPSEVVGRRWR--------VRA 151
A + G + R L V + R LT + L ++ P G W VRA
Sbjct 40 AALTPTGFRWDRNWLIVNSKGRGLTQRVEPKLSLIEVEMPKHAFGEDWEPEKSSNMVVRA 99
Query 152 DGSKLMKVHLDPKDKAKDNLEEKLWTFAAV 181
G +KV L DK D + W+ +A+
Sbjct 100 PGMDALKVSLAKPDKIADGVSVWEWSGSAL 129
Lambda K H
0.320 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3443493696
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40