bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0723_orf3
Length=135
Score E
Sequences producing significant alignments: (Bits) Value
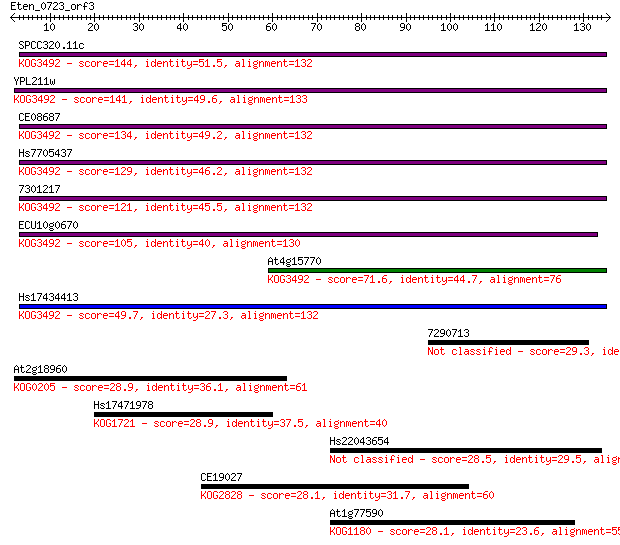
SPCC320.11c 144 5e-35
YPL211w 141 3e-34
CE08687 134 7e-32
Hs7705437 129 1e-30
7301217 121 3e-28
ECU10g0670 105 2e-23
At4g15770 71.6 4e-13
Hs17434413 49.7 2e-06
7290713 29.3 2.2
At2g18960 28.9 2.6
Hs17471978 28.9 3.1
Hs22043654 28.5 3.6
CE19027 28.1 4.6
At1g77590 28.1 5.3
> SPCC320.11c
Length=180
Score = 144 bits (363), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 68/132 (51%), Positives = 90/132 (68%), Gaps = 1/132 (0%)
Query 3 EKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAFV 62
E+ +K A V R+NL+S+G C GK TK KFRL ITAL IA+YA +K+W+ GE F+
Sbjct 49 ERAMKMATSVARQNLMSLGICFGKFTKTNKFRLHITALDYIAQYARYKIWVKSNGEMPFL 108
Query 63 YGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHA 122
YGNH++K HVGR+T D P GV + S+N D PLGFG +A++T E+ A+ +H A
Sbjct 109 YGNHVLKAHVGRITDDTPQHQGVVIYSMN-DTPLGFGVTARSTLELRRLEPTAIVAFHQA 167
Query 123 DVGEYLREEADL 134
DVGEYLR+E L
Sbjct 168 DVGEYLRDEDTL 179
> YPL211w
Length=181
Score = 141 bits (355), Expect = 3e-34, Method: Compositional matrix adjust.
Identities = 66/133 (49%), Positives = 93/133 (69%), Gaps = 1/133 (0%)
Query 2 PEKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAF 61
P+ + K A V R NL+S+G C+GK TK KFRL IT+L V+AK+A +K+W+ P GE F
Sbjct 48 PDHVAKLATSVARPNLMSLGICLGKFTKTGKFRLHITSLTVLAKHAKYKIWIKPNGEMPF 107
Query 62 VYGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHH 121
+YGNH++K HVG+++ D+P AGV V ++N D+PLGFG SAK+T+E + +
Sbjct 108 LYGNHVLKAHVGKMSDDIPEHAGVIVFAMN-DVPLGFGVSAKSTSESRNMQPTGIVAFRQ 166
Query 122 ADVGEYLREEADL 134
AD+GEYLR+E L
Sbjct 167 ADIGEYLRDEDTL 179
> CE08687
Length=180
Score = 134 bits (336), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 65/132 (49%), Positives = 86/132 (65%), Gaps = 1/132 (0%)
Query 3 EKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAFV 62
E L++ A C+ R+ LLS G C+GK TK +KF L ITAL +A YA KVWL P EQ F+
Sbjct 49 ENLMRQAACIAREPLLSFGTCLGKFTKSKKFHLQITALDYLAPYAKFKVWLKPNAEQQFL 108
Query 63 YGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHA 122
YGN+I+K + R+T P+ AG+ V S+ D+PLGFG SAK T++ A A+ H
Sbjct 109 YGNNILKSGIARMTDGTPTHAGIVVYSMT-DVPLGFGVSAKGTSDSKRADPTALVVLHQC 167
Query 123 DVGEYLREEADL 134
D+GEYLR E+ L
Sbjct 168 DLGEYLRNESHL 179
> Hs7705437
Length=180
Score = 129 bits (325), Expect = 1e-30, Method: Compositional matrix adjust.
Identities = 61/132 (46%), Positives = 86/132 (65%), Gaps = 1/132 (0%)
Query 3 EKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAFV 62
EK++K A + L+S+G C GK TK KFRL +TAL +A YA +KVW+ PG EQ+F+
Sbjct 49 EKIMKLAANISGDKLVSLGTCFGKFTKTHKFRLHVTALDYLAPYAKYKVWIKPGAEQSFL 108
Query 63 YGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHA 122
YGNH++K +GR+T + GV V S+ D+PLGFG +AK+T + A+ +H A
Sbjct 109 YGNHVLKSGLGRITENTSQYQGVVVYSM-ADIPLGFGVAAKSTQDCRKVDPMAIVVFHQA 167
Query 123 DVGEYLREEADL 134
D+GEY+R E L
Sbjct 168 DIGEYVRHEETL 179
> 7301217
Length=180
Score = 121 bits (304), Expect = 3e-28, Method: Compositional matrix adjust.
Identities = 60/132 (45%), Positives = 82/132 (62%), Gaps = 1/132 (0%)
Query 3 EKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAFV 62
E++LK + C K L+ VG C GK +K K + ITAL+ +A YA +KVW+ P EQ F+
Sbjct 49 ERILKLSECFGYKQLVCVGTCFGKFSKTNKLKFHITALYYLAPYAQYKVWVKPSFEQQFL 108
Query 63 YGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHA 122
YGNHI K +GR+T + GV V S+N DLPLGFG A++T + TA +H +
Sbjct 109 YGNHIPKTGLGRITENAGQYQGVVVYSMN-DLPLGFGVLARSTTDCKTADPMTTVCFHQS 167
Query 123 DVGEYLREEADL 134
D+GEY+R E L
Sbjct 168 DIGEYIRAEDTL 179
> ECU10g0670
Length=176
Score = 105 bits (263), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 52/130 (40%), Positives = 80/130 (61%), Gaps = 1/130 (0%)
Query 3 EKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAFV 62
E++ A + RKNL VG IGK TK FRLG+ +L++++++A HKVW+ E +V
Sbjct 46 ERMRAATSMISRKNLGMVGTVIGKFTKSGSFRLGVASLNLLSRWAIHKVWIKNSAEMNYV 105
Query 63 YGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHA 122
YGN+ +K HV R++ +P +GV V + + D+PLGFG +A + A A++ +
Sbjct 106 YGNNALKSHVFRISEGIPINSGVFVFNQH-DVPLGFGITALSPQGYSAAKGHALAILRQS 164
Query 123 DVGEYLREEA 132
D GEY+R EA
Sbjct 165 DTGEYVRNEA 174
> At4g15770
Length=75
Score = 71.6 bits (174), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 34/76 (44%), Positives = 49/76 (64%), Gaps = 1/76 (1%)
Query 59 QAFVYGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSF 118
+F+YGNH++K +GR+T + G GV V S++ D+PLGFG +AK+T + +
Sbjct 1 MSFLYGNHVLKGGLGRITDSIVPGDGVVVFSMS-DVPLGFGIAAKSTQDCRKLDPNGIVV 59
Query 119 YHHADVGEYLREEADL 134
H AD+GEYLR E DL
Sbjct 60 LHQADIGEYLRGEDDL 75
> Hs17434413
Length=133
Score = 49.7 bits (117), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 36/132 (27%), Positives = 50/132 (37%), Gaps = 48/132 (36%)
Query 3 EKLLKAAGCVPRKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQAFV 62
EK++K + L+S+G C GK+TK KFR
Sbjct 49 EKIVKLDANISGDKLVSLGTCFGKVTKTHKFRF--------------------------- 81
Query 63 YGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHA 122
GV V S+ D+PLGFG +AK+T + A+ H A
Sbjct 82 --------------------TGVVVYSM-ADIPLGFGVAAKSTQDCRKVDPMAIVVLHQA 120
Query 123 DVGEYLREEADL 134
D+GEY + E L
Sbjct 121 DIGEYAQHEETL 132
> 7290713
Length=436
Score = 29.3 bits (64), Expect = 2.2, Method: Composition-based stats.
Identities = 15/38 (39%), Positives = 25/38 (65%), Gaps = 2/38 (5%)
Query 95 PLGFGTSAKATA--EIHTAFTEAVSFYHHADVGEYLRE 130
PLG G S+KA + +IH+A+T+ + + VG+Y+ E
Sbjct 102 PLGSGRSSKAVSYQDIHSAYTKRRFQHVTSKVGQYIAE 139
> At2g18960
Length=949
Score = 28.9 bits (63), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 30/63 (47%), Gaps = 10/63 (15%)
Query 2 PEKLLKAAGCVP--RKNLLSVGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQ 59
PE++L A P RK +LS CI K + G+ +L V + P K +PGG
Sbjct 426 PEQILDLANARPDLRKKVLS---CIDKYAER-----GLRSLAVARQVVPEKTKESPGGPW 477
Query 60 AFV 62
FV
Sbjct 478 EFV 480
> Hs17471978
Length=714
Score = 28.9 bits (63), Expect = 3.1, Method: Composition-based stats.
Identities = 15/40 (37%), Positives = 20/40 (50%), Gaps = 0/40 (0%)
Query 20 VGQCIGKITKGRKFRLGITALHVIAKYAPHKVWLNPGGEQ 59
+G C GK +G+ AL V+A A L PGG+Q
Sbjct 431 LGSCAGKPGPTVGVFMGLGALRVVASQARQGTVLQPGGDQ 470
> Hs22043654
Length=239
Score = 28.5 bits (62), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query 73 GRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHADVGEYLREEA 132
G +T D+ +G G+ + N D L G KA I + E H++ G +L EE
Sbjct 106 GDVTGDLRNGVGLLTPTSNSDFSL--GAREKALEAIGCSKIEQTQLVLHSEDGAHLSEEK 163
Query 133 D 133
+
Sbjct 164 N 164
> CE19027
Length=177
Score = 28.1 bits (61), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 26/60 (43%), Gaps = 5/60 (8%)
Query 44 AKYAPHKVWLNPGGEQAFVYGNHIIKRHVGRLTADMPSGAGVCVLSLNGDLPLGFGTSAK 103
A Y P ++L E VY + + H +T P G C L ++ D L TSAK
Sbjct 70 ADYTP--IFL---SEVPSVYTSGTLNVHFALITVSPPDELGFCTLGVDIDTTLAAATSAK 124
> At1g77590
Length=691
Score = 28.1 bits (61), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 13/55 (23%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query 73 GRLTADMPSGAGVCVLSLNGDLPLGFGTSAKATAEIHTAFTEAVSFYHHADVGEY 127
G LT+D P G V+ ++ LG+ + + T E++ + + +++ D+G +
Sbjct 487 GYLTSDKPMPRGEIVIG-GSNITLGYFKNEEKTKEVYKVDEKGMRWFYTGDIGRF 540
Lambda K H
0.321 0.138 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1388795348
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40