bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0701_orf1
Length=198
Score E
Sequences producing significant alignments: (Bits) Value
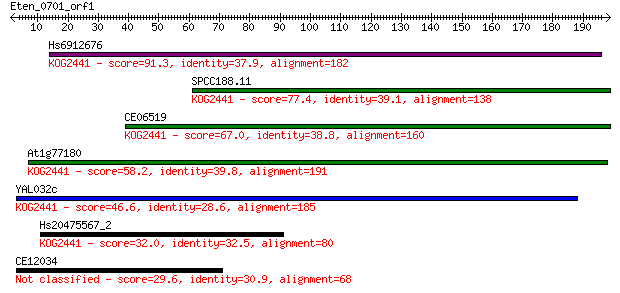
Hs6912676 91.3 1e-18
SPCC188.11 77.4 2e-14
CE06519 67.0 2e-11
At1g77180 58.2 1e-08
YAL032c 46.6 4e-05
Hs20475567_2 32.0 0.76
CE12034 29.6 4.6
> Hs6912676
Length=536
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 69/196 (35%), Positives = 107/196 (54%), Gaps = 25/196 (12%)
Query 14 LRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERSRGKR 73
LR++A +AR RA + + G+A E E + + R+ ++++ RA +RS+ +R
Sbjct 324 LREMAQKARERRAGIKTHVEKED-GEARERDEIRHDRRKERQHDRNLSRAAPDKRSKLQR 382
Query 74 RDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLF 133
++ RDISE++ALG+P + + +D RLFNQ G DSG+ GGED+ +YD+
Sbjct 383 NEN--RDISEVIALGVP--NPRTSNEVQYDQRLFNQSKGMDSGFAGGEDEIYNVYDQ--- 435
Query 134 AQRGGA----GIYQFSRDRFSTSVGEGSEL----------AAFAGADKTRYMRTGPVEFE 179
A RGG IY+ S++ G+ E F+G+D+ + R GPV+FE
Sbjct 436 AWRGGKDMAQSIYRPSKNLDKDMYGDDLEARIKTNRFVPDKEFSGSDRRQRGREGPVQFE 495
Query 180 KDVSDPFGLDNLLSEA 195
+ DPFGLD L EA
Sbjct 496 E---DPFGLDKFLEEA 508
> SPCC188.11
Length=557
Score = 77.4 bits (189), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 54/160 (33%), Positives = 80/160 (50%), Gaps = 29/160 (18%)
Query 61 ERAVRLERSRGKRR-----DDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTDS 115
E+ +RL R ++R D RD++E VALGL +D+M D+RLFNQ G S
Sbjct 403 EKDLRLSRMGAEKRAKLAEKDRPRDVAERVALGL--SKPSMSSDTMIDSRLFNQASGLGS 460
Query 116 GYKGGEDDTDTLYDRPLFA------QRGGAGIYQ-----------FSRDRFSTSVGEGSE 158
G++ ++D+ +YD+P A R GA + + S R+ G+
Sbjct 461 GFQ--DEDSYNVYDKPWRAAPSSTLYRPGATLSRQVDASAELERITSESRYDVL---GNA 515
Query 159 LAAFAGADKTRYMRTGPVEFEKDVSDPFGLDNLLSEAHKK 198
F G+D+ R GPV FEKD++DPFG+D L+ K
Sbjct 516 HKKFKGSDEVVESRAGPVTFEKDIADPFGVDTFLNNVSSK 555
> CE06519
Length=535
Score = 67.0 bits (162), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 62/186 (33%), Positives = 89/186 (47%), Gaps = 40/186 (21%)
Query 39 DAEEEAERQQREQIAREREKDIERAVRLERSRG----KRRDDSQRDISELVALGLPVDAK 94
D E++ + + RE+I R+R DI + + RSR K R + +RDISE + LGLP +
Sbjct 345 DDEDDEQVKVREEIRRDRLDDIRKERNIARSRPDKADKLRKERERDISEKIVLGLPDTNQ 404
Query 95 KQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLFAQRGGAGIYQF---------- 144
K+ + FD RLF++ G DSG +DDT YD A RGG + Q
Sbjct 405 KRTGEPQFDQRLFDKTQGLDSG--AMDDDTYNPYD---AAWRGGDSVQQHVYRPSKNLDN 459
Query 145 ------------SRDRFSTSVGEGSELAAFAGADKTRYMRTGPVEFEKDVSDPFGLDNLL 192
++RF G F+GA+ + +GPV+FEKD D FGL +L
Sbjct 460 DVYGGDLDKIIEQKNRFVADKG-------FSGAEGSSRG-SGPVQFEKD-QDVFGLSSLF 510
Query 193 SEAHKK 198
+K
Sbjct 511 EHTKEK 516
> At1g77180
Length=613
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 76/245 (31%), Positives = 105/245 (42%), Gaps = 58/245 (23%)
Query 7 EELREQQLRQLAAQARAERA----QLLQQSRRAAAGDAEEEAERQQREQ----------- 51
+E +EQ+LR LA +AR+ER + S R + + + +Q
Sbjct 328 KERKEQELRALAQKARSERTGAAMSMPVSSDRGRSESVDPRGDYDNYDQDRGREREREEP 387
Query 52 --IAREREKDIER---------------------AVRLERSRGKRRDDSQRDISELVALG 88
EREK I+R A ++S+ R D RDISE VALG
Sbjct 388 QETREEREKRIQREKIREERRRERERERRLDAKDAAMGKKSKITR--DRDRDISEKVALG 445
Query 89 LPVDAKKQRTDSMFDTRLFNQGGGTDSGYKGGEDDTDTLYDRPLF-AQRGGAGIYQFSRD 147
+ K + M+D RLFNQ G DSG+ DD LYD+ LF AQ + +Y+ +D
Sbjct 446 MASTGGKGGGEVMYDQRLFNQDKGMDSGF--AADDQYNLYDKGLFTAQPTLSTLYKPKKD 503
Query 148 RFSTSVGEGSEL-------------AAFAGA-DKTRYMRTGPVEFEK-DVSDPFGLDNLL 192
G E AF GA ++ R PVEFEK + DPFGL+ +
Sbjct 504 NDEEMYGNADEQLDKIKNTERFKPDKAFTGASERVGSKRDRPVEFEKEEEQDPFGLEKWV 563
Query 193 SEAHK 197
S+ K
Sbjct 564 SDLKK 568
> YAL032c
Length=379
Score = 46.6 bits (109), Expect = 4e-05, Method: Compositional matrix adjust.
Identities = 53/197 (26%), Positives = 90/197 (45%), Gaps = 43/197 (21%)
Query 3 KKSAEELREQ-QLRQLAAQ----ARAERAQLLQQSRRAAAGDAEEEA---ERQQREQIAR 54
KK+ +E+R + +L++LA + A+ + + L Q R G + A ++Q +AR
Sbjct 213 KKARQEIRSKMELKRLAMEQEMLAKESKLKELSQRARYHNGTPQTGAIVKPKKQTSTVAR 272
Query 55 EREKDIERAVRLERSRGKRRDDSQRDISELVALGLPVDAKKQRTDSMFDTRLFNQGGGTD 114
+E L S+G RD+SE + LG + ++ D +D+R F +G
Sbjct 273 LKE--------LAYSQG-------RDVSEKIILG--AAKRSEQPDLQYDSRFFTRGANA- 314
Query 115 SGYKGGEDDTDTLYDRPLFAQRGGAGIYQFSRDRFSTSVGEGSELAAFAGADKTRYMRTG 174
+ D +YD PLF Q+ IY+ + ++ +V SE GA + G
Sbjct 315 ----SAKRHEDQVYDNPLFVQQDIESIYKTNYEKLDEAVNVKSE-----GASGSH----G 361
Query 175 PVEFEK----DVSDPFG 187
P++F K D SD +G
Sbjct 362 PIQFTKAESDDKSDNYG 378
> Hs20475567_2
Length=320
Score = 32.0 bits (71), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 26/84 (30%), Positives = 45/84 (53%), Gaps = 11/84 (13%)
Query 11 EQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIERAVRLERSR 70
E++LR++A +AR RA + + E+ E ++R +I +R K+ + L R+
Sbjct 242 EEKLREMAQKARERRAGIKTHVEK-------EDREARERNEIRHDRRKERQHDQNLSRAA 294
Query 71 GKRRDDSQR----DISELVALGLP 90
+R QR DISE++AL +P
Sbjct 295 PDKRSKLQRNENGDISEVIALCVP 318
> CE12034
Length=1355
Score = 29.6 bits (65), Expect = 4.6, Method: Composition-based stats.
Identities = 21/68 (30%), Positives = 36/68 (52%), Gaps = 7/68 (10%)
Query 3 KKSAEELREQQLRQLAAQARAERAQLLQQSRRAAAGDAEEEAERQQREQIAREREKDIER 62
+++ ++ RE R AQ R ER Q L+ A D E+E R++ + ARE K ++
Sbjct 128 ERATQKEREHIERHREAQERHERLQALE------AKDKEDERAREEDKAKAREEAKS-KK 180
Query 63 AVRLERSR 70
+ E+S+
Sbjct 181 DTKAEKSK 188
Lambda K H
0.312 0.130 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3407623970
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40