bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0686_orf1
Length=346
Score E
Sequences producing significant alignments: (Bits) Value
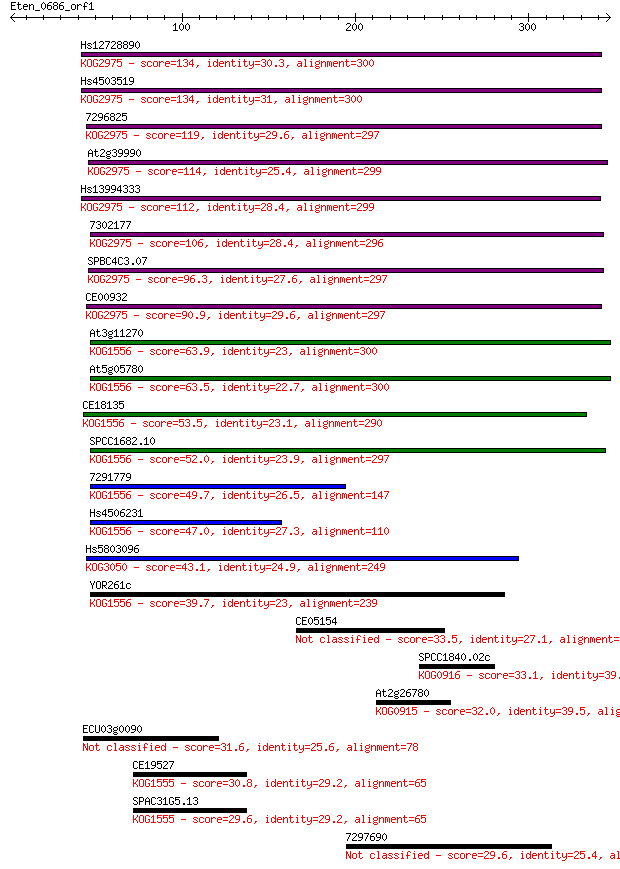
Hs12728890 134 3e-31
Hs4503519 134 4e-31
7296825 119 1e-26
At2g39990 114 3e-25
Hs13994333 112 2e-24
7302177 106 6e-23
SPBC4C3.07 96.3 1e-19
CE00932 90.9 3e-18
At3g11270 63.9 5e-10
At5g05780 63.5 6e-10
CE18135 53.5 7e-07
SPCC1682.10 52.0 2e-06
7291779 49.7 8e-06
Hs4506231 47.0 7e-05
Hs5803096 43.1 8e-04
YOR261c 39.7 0.009
CE05154 33.5 0.76
SPCC1840.02c 33.1 0.98
At2g26780 32.0 2.3
ECU03g0090 31.6 2.9
CE19527 30.8 5.0
SPAC31G5.13 29.6 8.9
7297690 29.6 8.9
> Hs12728890
Length=361
Score = 134 bits (337), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 91/302 (30%), Positives = 153/302 (50%), Gaps = 34/302 (11%)
Query 42 PFPSLRV-RLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSLTG 100
PFP RV RLHPV L +I+D+Y RR+EG VIGTLLG+ D + ++VT+CF H+
Sbjct 89 PFPGGRVVRLHPVILASIVDSYERRNEGAARVIGTLLGTV-DKHSVEVTNCFSVPHN-ES 146
Query 101 EGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVS 160
E + + + ++M+EL ++ + E+++GW+ TG ++TE + +H ++ + +
Sbjct 147 EDEVAVDMEFAKNMYELHKKVSP-----NELILGWYATGHDITEHSVLIHEYYSREAP-- 199
Query 161 KFFPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTM-AREACFQFHELPLELYATPSDR 219
PIHL VD +++G S+K Y M + R F L ++ ++R
Sbjct 200 ---------NPIHLTVDTSLQNGRMSIKAYVSTLMGVPGRTMGVMFTPLTVKYAYYDTER 250
Query 220 VGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRR 279
+G++L+ K P LL D+ + ++Q+ L Y
Sbjct 251 IGVELIMKT-------------CFSPNRVIGLLSDLQQ-VGGASARIQDALSTVLQYAED 296
Query 280 VLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQFAVAQ 339
VL+G+V D VGRFL S ++ + + FE + + + D LM +LA L + Q A+ +
Sbjct 297 VLSGKVSADNTVGRFLMSLVNRVPKIVPDDFETMLNSNINDLLMVTYLANLTQSQIALNE 356
Query 340 KL 341
KL
Sbjct 357 KL 358
> Hs4503519
Length=357
Score = 134 bits (336), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 93/309 (30%), Positives = 156/309 (50%), Gaps = 48/309 (15%)
Query 42 PFPSLRV-RLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSLTG 100
PFP RV RLHPV L +I+D+Y RR+EG VIGTLLG+ D + ++VT+CF H+
Sbjct 85 PFPGGRVVRLHPVILASIVDSYERRNEGAARVIGTLLGTV-DKHSVEVTNCFSVPHN-ES 142
Query 101 EGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVS 160
E + + + ++M+EL ++ + E+++GW+ TG ++TE + +H ++ + +
Sbjct 143 EDEVAVDMEFAKNMYELHKKVSP-----NELILGWYATGHDITEHSVLIHEYYSREAP-- 195
Query 161 KFFPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTM-AREACFQFHELPLELYATPSDR 219
PIHL VD +++G S+K Y M + R F L ++ ++R
Sbjct 196 ---------NPIHLTVDTSLQNGRMSIKAYVSTLMGVPGRTMGVMFTPLTVKYAYYDTER 246
Query 220 VGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQ-------LQEKLEI 272
+G+ L+ K + P R GL+++L+Q +Q+ L
Sbjct 247 IGVDLIMKTCFS----PNRVI-----------------GLSSDLQQVGGASARIQDALST 285
Query 273 CAAYVRRVLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAK 332
Y VL+G+V D VGRFL S ++ + + FE + + + D LM +LA L +
Sbjct 286 VLQYAEDVLSGKVSADNTVGRFLMSLVNQVPKIVPDDFETMLNSNINDLLMVTYLANLTQ 345
Query 333 LQFAVAQKL 341
Q A+ +KL
Sbjct 346 SQIALNEKL 354
> 7296825
Length=280
Score = 119 bits (298), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 88/302 (29%), Positives = 145/302 (48%), Gaps = 37/302 (12%)
Query 45 SLRVRLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFV---DRHSLTGE 101
+L VR+HPV L ++DA+ RR+ VIGTLLGS G +++VT+CF H E
Sbjct 5 NLTVRVHPVVLFQVVDAFERRNADSHRVIGTLLGSVDKG-VVEVTNCFCVPHKEHDDQVE 63
Query 102 GLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSK 161
L D M++L ++ N E VVGW+ TG+++T + +H ++ +
Sbjct 64 AELSYALD----MYDLNRKVNSN-----ESVVGWWATGNDVTNHSSVIHEYYAR------ 108
Query 162 FFPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTM--AREACFQFHELPLELYATPSDR 219
P+HL VD ++ G ++ Y +Q+ + + C F +P+EL + +
Sbjct 109 -----ECNNPVHLTVDTSLQGGRMGLRAYVCIQLGVPGGKSGCM-FTPIPVELTSYEPET 162
Query 220 VGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRR 279
GL+LLQK +H P+ T P +LD ++ +LQ L++ YV
Sbjct 163 FGLKLLQKTVGVSPAHRPK----TVPP----MLD--LAQISEASTKLQSLLDLILKYVDD 212
Query 280 VLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQFAVAQ 339
V+ +V PD VGR L + E F Q+ +++ L+ L++L K Q + +
Sbjct 213 VIAHKVTPDNAVGRQLLDLIHSVPHMTHEQFTQMFNANVRNLLLVITLSQLIKTQLQLNE 272
Query 340 KL 341
KL
Sbjct 273 KL 274
> At2g39990
Length=293
Score = 114 bits (286), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 76/300 (25%), Positives = 149/300 (49%), Gaps = 37/300 (12%)
Query 46 LRVRLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSLTGEGLLQ 105
L R+HP+ + + D ++RR + E VIGTLLGS +D+ + + H+ + + +
Sbjct 26 LTARIHPLVIFNVCDCFVRRPDSAERVIGTLLGSILPDGTVDIRNSYAVPHNESSDQVAV 85
Query 106 IIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSKFFPQ 165
I D+H +M + N +E +VGW+ TG+ + + +H ++ +
Sbjct 86 DI-DYHHNMLASHLKVNS-----KETIVGWYSTGAGVNGGSSLIHDFYAR---------- 129
Query 166 PPLTEPIHLMVDAVMESGSFSVKVYTQVQMTMA-REACFQFHELPLELYATPSDRVGLQL 224
+ PIHL VD +G ++K + +++ R+ F E+P++L ++RVG +
Sbjct 130 -EVPNPIHLTVDTGFTNGEGTIKAFVSSNLSLGDRQLVAHFQEIPVDLRMVDAERVGFDV 188
Query 225 LQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRRVLNGE 284
L+ S L +D+ +G+ +E+L + YV V+ G+
Sbjct 189 LKA------------------TSVDKLPNDL-EGMELTMERLLTLINDVYKYVDSVVGGQ 229
Query 285 VEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQFAVAQKLNTS 344
+ PD +GRF++ A++ + + F+ L ++LQD L+ +L+ + + Q ++A+KLNT+
Sbjct 230 IAPDNNIGRFIADAVASLPKLPPQVFDNLVNDSLQDQLLLLYLSSITRTQLSLAEKLNTA 289
> Hs13994333
Length=348
Score = 112 bits (279), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 85/312 (27%), Positives = 154/312 (49%), Gaps = 55/312 (17%)
Query 42 PFPSLRV-RLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSLTG 100
PFP RV RLHPV L + +D+Y RR++G VIGTLLG+ + + ++VT+CF ++ +
Sbjct 75 PFPGGRVVRLHPVLLASTVDSYERRNKGAARVIGTLLGTVNKHS-VEVTNCFSVSYNESD 133
Query 101 EGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVS 160
E ++ + + ++M+EL ++ + E+++GW+ G ++TE + +H ++ + +
Sbjct 134 EVVVDM--EFAKNMYELHKKVSPN-----ELILGWYAAGHDITEHSVLIHEYYSREAP-- 184
Query 161 KFFPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTMAREACFQFHELPLE--LYATPSD 218
PIHL VD +++G S+K Y M + F L ++ Y T
Sbjct 185 ---------NPIHLTVDTSLQNGHMSIKAYVSTLMGVPGRTVGVFTPLTVKYAYYDTECI 235
Query 219 RVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQ-------LQEKLE 271
RV L +++ + +R GL+++L+Q +Q+ L
Sbjct 236 RVDL-IMKTCFSPNR----------------------VVGLSSDLQQVGGASAHIQDALS 272
Query 272 ICAAYVRRVLNGEVEPDP---EVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLA 328
I Y +L+ +V D +VG FL S ++ + + FE + + + D LM A+LA
Sbjct 273 IVLQYAEDILSAKVSADNTIRKVGHFLMSLVNQVPKIVPDDFETMLHSNINDLLMVAYLA 332
Query 329 RLAKLQFAVAQK 340
L + Q A+ +K
Sbjct 333 NLTQSQIALNEK 344
> 7302177
Length=285
Score = 106 bits (265), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 84/304 (27%), Positives = 141/304 (46%), Gaps = 41/304 (13%)
Query 47 RVRLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSLTGEG-LLQ 105
+V L P+ I+DAY RR +G V+GTLLG +G+I ++T+CF H E +
Sbjct 10 KVYLKPLVFFQIIDAYDRRPKGDNQVMGTLLGRNKEGHI-EITNCFTVPHKEHSENKRID 68
Query 106 IIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSKFFPQ 165
+ + + EL A E V+GWFCTG ++ +H ++ +
Sbjct 69 LDMAYASEVLELNMFAYPN-----ERVLGWFCTGKSVSRSASLIHDYYVRECCEG----- 118
Query 166 PPLTEPIHLMVDAVMESGSFSVKVYTQVQMTM-AREACFQFHELPLELYATPSDRVGLQL 224
+P+HL+VDA +++ S ++Y V+M + F +PLE+ SD V L+
Sbjct 119 ----QPLHLLVDAALKNQRLSTRLYCAVEMGVPGGTKGLMFSLVPLEISNENSDLVALRC 174
Query 225 LQKVRTA------HRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVR 278
++K R P A+ ++D D +Q +L++ Y+
Sbjct 175 IEKQSQQQASKQMERFVPELAQ----------VVDATRD--------MQHRLDLVLRYIN 216
Query 279 RVLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQFAVA 338
VL + +PD VGR L +AL+ D + F + L+D LMA L+ + K Q ++
Sbjct 217 DVLARKKKPDNVVGRSLYAALTAVPLLDSDKFRVMFNTNLRDMLMAITLSTMIKTQLEIS 276
Query 339 QKLN 342
+KL+
Sbjct 277 EKLS 280
> SPBC4C3.07
Length=302
Score = 96.3 bits (238), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 82/308 (26%), Positives = 138/308 (44%), Gaps = 40/308 (12%)
Query 46 LRVRLHPVPLMTILDAYIRRDEGQENVIGTLLGSCS-DGNIIDVTDCFVDRHSLTGEGLL 104
L + + P L +ILD R+ E + VIGTLLG+ S DG I++ CF H+ + E +
Sbjct 21 LNIVIEPAVLFSILDHSTRKSENNQRVIGTLLGTRSEDGREIEIKSCFAVPHNESSEQV- 79
Query 105 QIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSKFFP 164
++ ++H +M+ L +AN REVVVGW+ T ++ + + + P
Sbjct 80 EVEMEYHRAMYHLHLKANP-----REVVVGWYATSPDLDAFSALIQNLYAS--------P 126
Query 165 QPPLTEP--------IHLMVDAVMESGSFSVKVY--TQVQMTMAREACFQFHELPLELYA 214
P T P +HL V+ + S ++K Y + V +T F P +
Sbjct 127 AEPGTAPLGTYPHPCVHLTVNTDV-SSPLAIKTYVSSPVGITERLADSCAFVPTPFTIRD 185
Query 215 TPSDRVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICA 274
+ R GL+ + A +P+ L D+ + LE L +E +
Sbjct 186 DEAVRSGLKAV-------------AAPKNDPSRLASLFTDLQQLRRSTLELLS-MIERVS 231
Query 275 AYVRRVLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQ 334
YV+ V++G + VGR+L S + + FE++ + LQD L+ +LA + Q
Sbjct 232 DYVQNVIDGSSPANVAVGRYLMKCFSLIPCVEGQDFEKIFSSHLQDVLVVVYLANTLRTQ 291
Query 335 FAVAQKLN 342
+A +LN
Sbjct 292 VDIASRLN 299
> CE00932
Length=294
Score = 90.9 bits (224), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 88/315 (27%), Positives = 145/315 (46%), Gaps = 45/315 (14%)
Query 45 SLRVRLHPVPLMTILDAYIRR------DEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSL 98
+L V +HP M ++D ++RR + GQE +GTL+G G+I VT+CF +
Sbjct 4 NLTVNVHPGVYMNVVDTHMRRTKSSAKNTGQEKCMGTLMGYYEKGSI-QVTNCFAIPFNE 62
Query 99 TGEGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWF-KQFS 157
+ + L +I ++ M ++ + E VGWF T S++T H ++ + +
Sbjct 63 SNDDL-EIDDQFNQQMISALKKTSPN-----EQPVGWFLTTSDITSSCLIYHDYYVRVIT 116
Query 158 SVSKFFPQPPLTEPIHLMVDAVMESGSFS----VKVYTQVQMTMAREA---CFQFHELPL 210
S PP+ + L +D SG S V+ Y + + + A C F+ L +
Sbjct 117 EASARRESPPI---VVLTIDTTF-SGDMSKRMPVRAYLRSKAGIPGAAGPHCAIFNPLRV 172
Query 211 ELYATPSDRVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKL 270
EL A P + V +QL++K + R L+ + L Q+ E L
Sbjct 173 ELAAFPGELVAMQLIEKALDSRRRE--------------ATLESGLEQLETSTAQMIEWL 218
Query 271 EICAAYVRRV-LNGEVEPDPEVGRFLS---SALSGATEPDLEAFEQLCQNTLQDSLMAAH 326
E YV V NGE D ++GR L +A S +P E + L +NTL+D +M ++
Sbjct 219 ERMLHYVEDVNKNGEKPGDAQIGRQLMDIVTASSNNMQP--EKLDTLVKNTLRDYVMVSY 276
Query 327 LARLAKLQFAVAQKL 341
LA+L + Q V ++L
Sbjct 277 LAKLTQTQLQVHERL 291
> At3g11270
Length=310
Score = 63.9 bits (154), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 69/311 (22%), Positives = 135/311 (43%), Gaps = 48/311 (15%)
Query 47 RVRLHPVPLMTILDAYIR-RDEGQENVIGTLLGSCSDGNIIDVTDCFV------DRHSLT 99
+V +HP+ L++I+D Y R + + V+G LLGS S G + DVT+ + D+ +
Sbjct 16 KVIVHPLVLLSIVDHYNRVAKDTSKRVVGVLLGSSSRGTV-DVTNSYAVPFEEDDKDT-- 72
Query 100 GEGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSV 159
+ + ++HESMF + ++ N +E +VGW+ TG ++ E VH F
Sbjct 73 --SIWFLDHNYHESMFHMFKRINA-----KEHIVGWYSTGPKLRENDLDVHALF------ 119
Query 160 SKFFPQPPLTEPIHLMVDAVMESGSFSVKVY---TQVQMTMAREACFQFHELPLELYATP 216
+ + P P L +++D + K Y +V+ +++ F +P E+ A
Sbjct 120 NGYVPNPVL-----VIIDVQPKELGIPTKAYYAVEEVKENATQKSQQVFVHVPTEIAAHE 174
Query 217 SDRVGLQ-LLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAA 275
+ +G++ LL+ V+ S L ++T L A L+ L +L
Sbjct 175 VEEIGVEHLLRDVKDTTIS---------------TLATEVTAKLTA-LKGLDARLREIRT 218
Query 276 YVRRVLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQF 335
Y+ V+ G++ + E+ L + ++ + D ++ +L+ L +
Sbjct 219 YLDLVIEGKLPLNHEILYHLQDVFNLLPNLNVNELVKAFAVKTNDMMLVIYLSSLIRSVI 278
Query 336 AVAQKLNTSFF 346
A+ +N
Sbjct 279 ALHSLINNKLL 289
> At5g05780
Length=308
Score = 63.5 bits (153), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 68/307 (22%), Positives = 133/307 (43%), Gaps = 40/307 (13%)
Query 47 RVRLHPVPLMTILDAYIR-RDEGQENVIGTLLGSCSDGNIIDVTDCFVD--RHSLTGEGL 103
+V +HP+ L++I+D Y R + + V+G LLGS S G ++DVT+ + +
Sbjct 16 KVVVHPLVLLSIVDHYNRVAKDSSKRVVGVLLGSSSRG-VVDVTNSYAVPFEEDDKDPSI 74
Query 104 LQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSKFF 163
+ ++HESMF + ++ N +E VVGW+ TG ++ E VH F + +
Sbjct 75 WFLDHNYHESMFHMFKRINA-----KEHVVGWYSTGPKLRENDLDVHALF------NGYV 123
Query 164 PQPPLTEPIHLMVDAVMESGSFSVKVY---TQVQMTMAREACFQFHELPLELYATPSDRV 220
P P L +++D + K Y +V+ +++ F + E+ A + +
Sbjct 124 PNPVL-----VIIDVQPKELGIPTKAYYAVEEVKENATQKSQKVFVHVSTEIAAHEVEEI 178
Query 221 GLQ-LLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRR 279
G++ LL+ V+ S L ++T L A L+ L +L +Y+
Sbjct 179 GVEHLLRDVKDTTIS---------------TLATEVTAKLTA-LKGLDARLREIRSYLDL 222
Query 280 VLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQLCQNTLQDSLMAAHLARLAKLQFAVAQ 339
V+ G++ + E+ L + ++ + D ++ +L+ L + A+
Sbjct 223 VIEGKLPLNHEILYHLQDVFNLLPNLNVNELVKAFSVKTNDMMLVIYLSSLIRSVIALHN 282
Query 340 KLNTSFF 346
+N
Sbjct 283 LINNKLL 289
> CE18135
Length=362
Score = 53.5 bits (127), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 67/304 (22%), Positives = 125/304 (41%), Gaps = 53/304 (17%)
Query 43 FPSLRVRLHPVPLMTILDAYIRRDEGQ--ENVIGTLLGSCSDGNIIDVTDCFV------D 94
P +V +HP+ L++++D + R + Q + V+G LLGS +D+ + F D
Sbjct 36 LPVNKVTVHPLVLLSVVDHFNRVSKTQSVKRVVGVLLGSMKKDKTLDIGNSFAVPFDEDD 95
Query 95 RHSLTGEGLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFK 154
+ T L + D+ ESM+ + + +E +VGW+ TG ++ + A++ K
Sbjct 96 KDKSTW--FLDM--DYLESMYGMFYKV-----AAKEKIVGWYHTGPKLHKNDIAINEQLK 146
Query 155 QFSSVSKFFPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTM--AREACFQFHELPLEL 212
+F P P L +++DA ++ + Y +VQ F +P ++
Sbjct 147 ------RFCPNPVL-----VIIDAEPKNIGLPTEAYIEVQEVHDDGTPPIKTFEHVPSDI 195
Query 213 -YATPSDRVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLE 271
+ LL+ ++ + G L ITD L L LQ +LE
Sbjct 196 GAEEAEEVGVEHLLRDIKD---------------QTAGTLSQRITDQLMG-LRGLQSQLE 239
Query 272 ICAAYVRRVLNGEVEPDPEVGRFLSSALS---GATEPDLEAFEQLCQNTLQDSLMAAHLA 328
Y+ ++ G + + V ++ L+ T PD + + N D LM ++
Sbjct 240 SIEKYLHDIVRGTLPVNHHVIYYVQEVLNLLPDVTHPDYIVSQNVQTN---DQLMCVYMG 296
Query 329 RLAK 332
L +
Sbjct 297 SLVR 300
> SPCC1682.10
Length=324
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 71/313 (22%), Positives = 139/313 (44%), Gaps = 49/313 (15%)
Query 47 RVRLHPVPLMTILDAYIRRDEG-QENVIGTLLGSCSDGNIIDVTDCFVDRHSLTGEGLLQ 105
+V +HP+ L++ +D+Y R +G + V+G LLG ++G++++V + + +
Sbjct 16 QVIVHPLVLLSAVDSYNRSAKGTKRRVVGILLGQ-NNGDVVNVANSYAIPFEEDEKNASV 74
Query 106 IIKDHH--ESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSKFF 163
DH+ ESM E+ ++ N E +VGW+ TG ++ ++ K K+
Sbjct 75 WFLDHNFMESMNEMFKKINAN-----EKLVGWYHTGPQLRPSDLEINNLLK------KYI 123
Query 164 PQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTM--AREACFQFHELPLELYATPSDRVG 221
P P L +++D +S Y + ++ F LP + A ++ +G
Sbjct 124 PNPVL-----VIIDVKPKSVGLPTNAYFAIDEIEDDGSKSSRTFVHLPSSIEAEEAEEIG 178
Query 222 LQ-LLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRRV 280
++ LL+ R AS G L +T + L+ L ++L A Y+R+V
Sbjct 179 VEHLLRDTRD---------------ASVGTLATRVTQQAQS-LQGLGQRLTEIADYLRKV 222
Query 281 LNGEVEPD-------PEVGRFLSSALSG--ATEPDLEAFEQLCQN-TLQDSLMAAHLARL 330
++G++ + V L + SG +E LE+ Q N D LM+ +++ +
Sbjct 223 VDGQLPINHAILAELQSVFNLLPNIFSGPVVSEQALESEAQRAFNVNSNDQLMSIYISSI 282
Query 331 AKLQFAVAQKLNT 343
+ A+ L++
Sbjct 283 VRAVIALHDLLDS 295
> 7291779
Length=338
Score = 49.7 bits (117), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 39/158 (24%), Positives = 79/158 (50%), Gaps = 17/158 (10%)
Query 47 RVRLHPVPLMTILDAYIRRDE--GQENVIGTLLGSCSDGNIIDVTDCFV---DRHSLTGE 101
+V +HP+ L++++D + R + Q+ V+G LLG ++DV++ F D +
Sbjct 10 KVIVHPLVLLSVVDHFNRMGKIGNQKRVVGVLLGCWRSKGVLDVSNSFAVPFDEDD-KDK 68
Query 102 GLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQF--SSV 159
+ + D+ E+M+ + ++ N RE VVGW+ TG ++ + A++ +++ +SV
Sbjct 69 SVWFLDHDYLENMYGMFKKVNA-----RERVVGWYHTGPKLHQNDIAINELVRRYCPNSV 123
Query 160 SKFFPQPP----LTEPIHLMVDAVMESGSFSVKVYTQV 193
P L ++ V+ V + GS + K + V
Sbjct 124 LVIIDAKPKDLGLPTEAYISVEEVHDDGSPTSKTFEHV 161
> Hs4506231
Length=324
Score = 47.0 bits (110), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 30/115 (26%), Positives = 63/115 (54%), Gaps = 12/115 (10%)
Query 47 RVRLHPVPLMTILDAYIR--RDEGQENVIGTLLGSCSDGNIIDVTDCFV---DRHSLTGE 101
+V +HP+ L++++D + R + Q+ V+G LLGS ++DV++ F D +
Sbjct 8 KVVVHPLVLLSVVDHFNRIVKVGNQKRVVGVLLGSWQ-KKVLDVSNSFAVPFDEDD-KDD 65
Query 102 GLLQIIKDHHESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQF 156
+ + D+ E+M+ + ++ N RE +VGW+ TG ++ + A++ K++
Sbjct 66 SVWFLDHDYLENMYGMFKKVNA-----RERIVGWYHTGPKLHKNDIAINELMKRY 115
> Hs5803096
Length=297
Score = 43.1 bits (100), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 62/254 (24%), Positives = 109/254 (42%), Gaps = 38/254 (14%)
Query 45 SLRVRLHPVPLMTILDAYI--RRDEGQE-NVIGTLLGSCSDGNIIDVTDCFVDRHSLTGE 101
S+ V LHP+ ++ I D +I R EG+ VIG L+G +G I+V + F + S T E
Sbjct 8 SVSVALHPLVILNISDHWIRMRSQEGRPVQVIGALIGK-QEGRNIEVMNSF-ELLSHTVE 65
Query 102 GLLQIIKDHHESMFELKQQANGGNGGVREV-VVGWFCTGSEMTELTCAVHGWFKQFSSVS 160
+ I K+++ + E +Q +E+ +GW+ TG VH KQ +
Sbjct 66 EKIIIDKEYYYTKEEQFKQV------FKELEFLGWYTTGGPPDPSDIHVH---KQVCEI- 115
Query 161 KFFPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTMAREACFQFHELPLELYATPSDRV 220
+ P+ L ++ + + V V+ V + EA F EL L ++R+
Sbjct 116 -------IESPLFLKLNPMTKHTDLPVSVFESVIDIINGEATMLFAELTYTLATEEAERI 168
Query 221 GLQLLQKVR-TAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRR 279
G+ + ++ T + AE L S ++ L ++++ YV+
Sbjct 169 GVDHVARMTATGSGENSTVAEHLIAQHS--------------AIKMLHSRVKLILEYVKA 214
Query 280 VLNGEVEPDPEVGR 293
GEV + E+ R
Sbjct 215 SEAGEVPFNHEILR 228
> YOR261c
Length=338
Score = 39.7 bits (91), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 55/246 (22%), Positives = 112/246 (45%), Gaps = 40/246 (16%)
Query 47 RVRLHPVPLMTILDAYIRRDEGQEN--VIGTLLGSCSDGNIIDVTDCFVDRHSLTGEGLL 104
+V + P+ L++ LD Y R + +EN +G +LG ++ + I VT+ F +
Sbjct 7 KVTIAPLVLLSALDHY-ERTQTKENKRCVGVILGD-ANSSTIRVTNSFALPFEEDEKNSD 64
Query 105 QIIKDHH--ESMFELKQQANGGNGGVREVVVGWFCTGSEMTELTCAVHGWFKQFSSVSKF 162
DH+ E+M E+ ++ N +E ++GW+ +G ++ ++ FK+++
Sbjct 65 VWFLDHNYIENMNEMCKKINA-----KEKLIGWYHSGPKLRASDLKINELFKKYTQ---- 115
Query 163 FPQPPLTEPIHLMVDAVMESGSFSVKVYTQVQMTMAREACFQ--FHELPLELYATPSDRV 220
P+ L+VD + Y ++ + F LP + A ++ +
Sbjct 116 ------NNPLLLIVDVKQQGVGLPTDAYVAIEQVKDDGTSTEKTFLHLPCTIEAEEAEEI 169
Query 221 GLQ-LLQKVRTAHRSHPPRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRR 279
G++ LL+ VR + A+ G+ + +T+ L + L+ LQ KL+ Y+ +
Sbjct 170 GVEHLLRDVR--------------DQAAGGLSI-RLTNQLKS-LKGLQSKLKDVVEYLDK 213
Query 280 VLNGEV 285
V+N E+
Sbjct 214 VINKEL 219
> CE05154
Length=552
Score = 33.5 bits (75), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 23/86 (26%), Positives = 40/86 (46%), Gaps = 5/86 (5%)
Query 166 PPLTEPIHLMVDAVMESGSFSV-KVYTQVQMTMAREACFQFHELPLELYATPSDRVGLQL 224
P + + MV+A+ + G S+ +V T+ + M +E E P+E++A P DR +
Sbjct 282 PAVESRMDAMVNALADGGRISLAEVRTRADVAMEQEQ----EEDPVEVFANPCDRQAVIY 337
Query 225 LQKVRTAHRSHPPRAEDLTEPASEGV 250
++ R P E+ E E V
Sbjct 338 KKRARVQTTDEPMETEEFFEVGIEDV 363
> SPCC1840.02c
Length=1955
Score = 33.1 bits (74), Expect = 0.98, Method: Compositional matrix adjust.
Identities = 17/43 (39%), Positives = 24/43 (55%), Gaps = 1/43 (2%)
Query 237 PRAEDLTEPASEGVLLDDITDGLNAELEQLQEKLEICAAYVRR 279
P A+++TEP EG LD+I L + Q ++ I YVRR
Sbjct 403 PEAQNVTEPVPEGYYLDNIVSPLYQYMHDQQFEI-INGKYVRR 444
> At2g26780
Length=1732
Score = 32.0 bits (71), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 17/44 (38%), Positives = 24/44 (54%), Gaps = 1/44 (2%)
Query 212 LYATPS-DRVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDD 254
LY S DRVG+ LL K+ ++ R E+L + + G LDD
Sbjct 245 LYIAASVDRVGMDLLVKIHSSQEPVAKRGEELLKKIASGTNLDD 288
> ECU03g0090
Length=622
Score = 31.6 bits (70), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 20/78 (25%), Positives = 38/78 (48%), Gaps = 1/78 (1%)
Query 43 FPSLRVRLHPVPLMTILDAYIRRDEGQENVIGTLLGSCSDGNIIDVTDCFVDRHSLTGEG 102
+ R RL + L+ +++ R + G N G L ++ I +++ D E
Sbjct 148 YKRARGRL-GIRLIDLVNRTFRHNIGMLNRFGQRLAQEAEAKIQEISSSLSDEEKRKEEK 206
Query 103 LLQIIKDHHESMFELKQQ 120
+LQIIK++ ES+ ++Q
Sbjct 207 MLQIIKEYGESLCTKEKQ 224
> CE19527
Length=312
Score = 30.8 bits (68), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 30/65 (46%), Gaps = 5/65 (7%)
Query 72 VIGTLLGSCSDGNIIDVTDCFVDRHSLTGEGLLQIIKDHHESMFELKQQANGGNGGVREV 131
V+G +LG D ++V D F S TG + + M ++ +Q G E+
Sbjct 55 VMGLMLGEFVDDYTVNVIDVFAMPQSGTGVSVEAVDPVFQAKMLDMLKQT-----GRPEM 109
Query 132 VVGWF 136
VVGW+
Sbjct 110 VVGWY 114
> SPAC31G5.13
Length=308
Score = 29.6 bits (65), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 31/65 (47%), Gaps = 5/65 (7%)
Query 72 VIGTLLGSCSDGNIIDVTDCFVDRHSLTGEGLLQIIKDHHESMFELKQQANGGNGGVREV 131
V+G +LG D + V D F S TG + + ++M ++ +Q G E+
Sbjct 52 VMGLMLGEFVDDFTVRVVDVFAMPQSGTGVSVEAVDPVFQKNMMDMLKQT-----GRPEM 106
Query 132 VVGWF 136
VVGW+
Sbjct 107 VVGWY 111
> 7297690
Length=717
Score = 29.6 bits (65), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 45/118 (38%), Gaps = 6/118 (5%)
Query 195 MTMAREACFQFHELPLELYATPSDRVGLQLLQKVRTAHRSHPPRAEDLTEPASEGVLLDD 254
M M R F Y PS++ L K H H P+ + TE S +L D
Sbjct 587 MDMLRFHKFTIGHAWCSDYGNPSEKEHFDNLYKFSPLHNVHTPKGAE-TEYPSTLILTAD 645
Query 255 ITDGLNAELEQLQEKLEICAAYVRRVLNGEVEPDPEVGRFLSSALSGATEPDLEAFEQ 312
D ++ L+ AA V + E + +P + R A GA +P + E+
Sbjct 646 HDDRVSP-----LHSLKFIAALQEAVRDSEFQKNPVLLRVYQKAGHGAGKPTSKRIEE 698
Lambda K H
0.321 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8340552822
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40