bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0675_orf2
Length=146
Score E
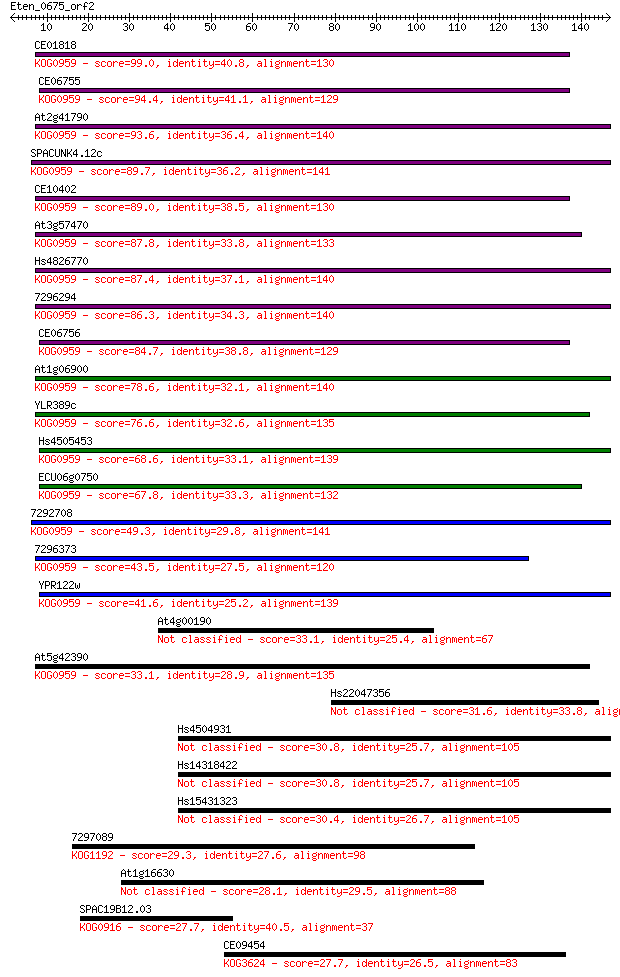
Sequences producing significant alignments: (Bits) Value
CE01818 99.0 3e-21
CE06755 94.4 8e-20
At2g41790 93.6 1e-19
SPACUNK4.12c 89.7 2e-18
CE10402 89.0 3e-18
At3g57470 87.8 6e-18
Hs4826770 87.4 7e-18
7296294 86.3 2e-17
CE06756 84.7 6e-17
At1g06900 78.6 3e-15
YLR389c 76.6 2e-14
Hs4505453 68.6 4e-12
ECU06g0750 67.8 7e-12
7292708 49.3 2e-06
7296373 43.5 1e-04
YPR122w 41.6 6e-04
At4g00190 33.1 0.20
At5g42390 33.1 0.22
Hs22047356 31.6 0.53
Hs4504931 30.8 0.87
Hs14318422 30.8 0.87
Hs15431323 30.4 1.2
7297089 29.3 2.7
At1g16630 28.1 6.2
SPAC19B12.03 27.7 7.2
CE09454 27.7 7.2
> CE01818
Length=745
Score = 99.0 bits (245), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 53/131 (40%), Positives = 78/131 (59%), Gaps = 1/131 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NAYT T +SF V + L L+ FFL+P+F+ + RE AV+ E+L K+N+D
Sbjct 101 SNAYTDTDHTNYSFEVRSEKLYGALDRFAQFFLDPQFTESATEREVCAVNCEYLDKVNED 160
Query 67 ALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
R + R SK +S F++GN +L+ +PR KGI+ + F+ WYSS++MT
Sbjct 161 FWRCLQVERSLSKPGHDYSKFAIGNKKTLLEDPRTKGIEPRDVLLDFYKNWYSSDIMTCC 220
Query 126 IVGAESLDRLQ 136
IVG ESLD L+
Sbjct 221 IVGKESLDVLE 231
> CE06755
Length=980
Score = 94.4 bits (233), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 53/130 (40%), Positives = 73/130 (56%), Gaps = 1/130 (0%)
Query 8 NAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKDA 67
NA T T + F V L L+ FFL+P+F+ + RE AV +E + LN D
Sbjct 101 NAVTASDHTNYHFDVKPDQLRGALDRFVQFFLSPQFTESATEREVCAVDSEHSNNLNNDL 160
Query 68 LRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAVI 126
RLS + R SK ++ F GN +L+ E RKKG++ + +F+ KWYSSN+MT I
Sbjct 161 WRLSQVDRSLSKPGHDYAKFGTGNKKTLLEEARKKGVEPRDALLQFYKKWYSSNIMTCCI 220
Query 127 VGAESLDRLQ 136
+G ESLD LQ
Sbjct 221 IGKESLDVLQ 230
> At2g41790
Length=970
Score = 93.6 bits (231), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 51/141 (36%), Positives = 77/141 (54%), Gaps = 1/141 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
NAYT T + F V A + L+ FF+ P SA + RE AV +E L D
Sbjct 99 TNAYTASEETNYHFDVNADCFDEALDRFAQFFIKPLMSADATMREIKAVDSENQKNLLSD 158
Query 67 ALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
R+ L + SK P+ FS GN+D+L P+ KG+D E+ +F+++ YS+N+M V
Sbjct 159 GWRIRQLQKHLSKEDHPYHKFSTGNMDTLHVRPQAKGVDTRSELIKFYEEHYSANIMHLV 218
Query 126 IVGAESLDRLQEMVVNTLGKI 146
+ G ESLD++Q++V +I
Sbjct 219 VYGKESLDKIQDLVERMFQEI 239
> SPACUNK4.12c
Length=969
Score = 89.7 bits (221), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 51/142 (35%), Positives = 79/142 (55%), Gaps = 1/142 (0%)
Query 6 LANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNK 65
++NAYT +T + F V AL L+ FF++P F + +RE AV +E L
Sbjct 97 ISNAYTASNNTNYYFEVSHDALYGALDRFAQFFIDPLFLEECKDREIRAVDSEHCKNLQS 156
Query 66 DALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTA 124
D+ R L S K+ FS F+ GN+++L P++ G+D +E+ +F+DK+YS+N+M
Sbjct 157 DSWRFWRLYSVLSNPKSVFSKFNTGNIETLGDVPKELGLDVRQELLKFYDKYYSANIMKL 216
Query 125 VIVGAESLDRLQEMVVNTLGKI 146
VI+G E LD LQ+ I
Sbjct 217 VIIGREPLDVLQDWAAELFSPI 238
> CE10402
Length=1067
Score = 89.0 bits (219), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 50/131 (38%), Positives = 72/131 (54%), Gaps = 1/131 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NAYT T + F V L L+ FFL+P+F+ + RE AV +E + LN D
Sbjct 159 SNAYTSSDHTNYHFDVKPDQLPGALDRFVQFFLSPQFTESATEREVCAVDSEHSNNLNND 218
Query 67 ALRLSYLIR-QSKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
R + R +SK + F GN +L+ + RKKGI+ + +FH KWYSS++MT
Sbjct 219 LWRFLQVDRSRSKPGHDYGKFGTGNKQTLLEDARKKGIEPRDALLQFHKKWYSSDIMTCC 278
Query 126 IVGAESLDRLQ 136
IVG E L+ L+
Sbjct 279 IVGKEPLNVLE 289
> At3g57470
Length=989
Score = 87.8 bits (216), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 45/134 (33%), Positives = 74/134 (55%), Gaps = 1/134 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
NAYT T + F + + + L+ FF+ P S + RE AV +E + L D
Sbjct 101 TNAYTSSEDTNYHFDINTDSFYEALDRFAQFFIQPLMSTDATMREIKAVDSEHQNNLLSD 160
Query 67 ALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
+ R++ L + S+ P+ FS GN+D+L P + G+D E+ +F+D+ YS+N+M V
Sbjct 161 SWRMAQLQKHLSREDHPYHKFSTGNMDTLHVRPEENGVDTRSELIKFYDEHYSANIMHLV 220
Query 126 IVGAESLDRLQEMV 139
+ G E+LD+ Q +V
Sbjct 221 VYGKENLDKTQGLV 234
> Hs4826770
Length=1019
Score = 87.4 bits (215), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 52/141 (36%), Positives = 73/141 (51%), Gaps = 1/141 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NA+T T + F V L L+ FFL P F +RE AV +E + D
Sbjct 138 SNAFTSGEHTNYYFDVSHEHLEGALDRFAQFFLCPLFDESCKDREVNAVDSEHEKNVMND 197
Query 67 ALRLSYLIRQS-KLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
A RL L + + K PFS F GN +L P ++GID +E+ +FH +YSSNLM
Sbjct 198 AWRLFQLEKATGNPKHPFSKFGTGNKYTLETRPNQEGIDVRQELLKFHSAYYSSNLMAVC 257
Query 126 IVGAESLDRLQEMVVNTLGKI 146
++G ESLD L +VV ++
Sbjct 258 VLGRESLDDLTNLVVKLFSEV 278
> 7296294
Length=990
Score = 86.3 bits (212), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 48/141 (34%), Positives = 72/141 (51%), Gaps = 1/141 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NA T T + F V L L+ FF+ P F+ + RE AV++E L D
Sbjct 111 SNAATYPLMTKYHFHVAPDKLDGALDRFAQFFIAPLFTPSATEREINAVNSEHEKNLPSD 170
Query 67 ALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
R+ + R +K +S F GN +L P+ K ID E+ +FH +WYS+N+M
Sbjct 171 LWRIKQVNRHLAKPDHAYSKFGSGNKTTLSEIPKSKNIDVRDELLKFHKQWYSANIMCLA 230
Query 126 IVGAESLDRLQEMVVNTLGKI 146
++G ESLD L+ MV+ +I
Sbjct 231 VIGKESLDELEGMVLEKFSEI 251
> CE06756
Length=845
Score = 84.7 bits (208), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 50/130 (38%), Positives = 68/130 (52%), Gaps = 1/130 (0%)
Query 8 NAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKDA 67
NA T T + F V L L+ FFL P+F+ + RE AV +E L LN D
Sbjct 101 NACTEPDHTYYHFDVKPDQLYGALDRFVQFFLCPQFTKSATEREVCAVDSEHLSNLNSDY 160
Query 68 LRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAVI 126
R+ + R S+ F GN +L+ + RKKGI+ + F+ KWYSSN+MT I
Sbjct 161 WRILQVDRSLSRPGHDNRKFCTGNKKTLLEDARKKGIEPRDALLEFYKKWYSSNIMTCCI 220
Query 127 VGAESLDRLQ 136
+G ESLD L+
Sbjct 221 IGKESLDVLE 230
> At1g06900
Length=1023
Score = 78.6 bits (192), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 45/141 (31%), Positives = 68/141 (48%), Gaps = 1/141 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NAYT T + F V L L FF+ P + RE LAV +EF L D
Sbjct 159 SNAYTEMEHTCYHFEVKREFLQGALKRFSQFFVAPLMKTEAMEREVLAVDSEFNQALQND 218
Query 67 ALRLSYLIRQSKLKT-PFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
A RL L + K PF+ F+ G +++ + G+D + + + + ++Y LM V
Sbjct 219 ACRLQQLQCYTSAKGHPFNRFAWGKDTAVLSGAMENGVDLRECIVKLYKEYYHGGLMKLV 278
Query 126 IVGAESLDRLQEMVVNTLGKI 146
++G ESLD L+ VV G +
Sbjct 279 VIGGESLDMLESWVVELFGDV 299
> YLR389c
Length=988
Score = 76.6 bits (187), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 44/136 (32%), Positives = 72/136 (52%), Gaps = 1/136 (0%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NAYT +T + F V L L+ FF P F+ +++E AV++E L D
Sbjct 148 SNAYTASQNTNYFFEVNHQHLFGALDRFSGFFSCPLFNKDSTDKEINAVNSENKKNLQND 207
Query 67 ALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAV 125
R+ L + + K P+ FS GN+++L P++ G++ E+ +FH +YS+NLM
Sbjct 208 IWRIYQLDKSLTNTKHPYHKFSTGNIETLGTLPKENGLNVRDELLKFHKNFYSANLMKLC 267
Query 126 IVGAESLDRLQEMVVN 141
I+G E LD L + +
Sbjct 268 ILGREDLDTLSDWTYD 283
> Hs4505453
Length=1219
Score = 68.6 bits (166), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 46/140 (32%), Positives = 66/140 (47%), Gaps = 1/140 (0%)
Query 8 NAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKDA 67
NA T TVF F V + L+ FF++P +RE AV +E+ DA
Sbjct 332 NASTDCERTVFQFDVQRKYFKEALDRWAQFFIHPLMIRDAIDREVEAVDSEYQLARPSDA 391
Query 68 LRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAVI 126
R L ++ P F GN ++L EPRK ID +R F ++YSS+ MT V+
Sbjct 392 NRKEMLFGSLARPGHPMGKFFWGNAETLKHEPRKNNIDTHARLREFWMRYYSSHYMTLVV 451
Query 127 VGAESLDRLQEMVVNTLGKI 146
E+LD L++ V +I
Sbjct 452 QSKETLDTLEKWVTEIFSQI 471
> ECU06g0750
Length=882
Score = 67.8 bits (164), Expect = 7e-12, Method: Composition-based stats.
Identities = 44/133 (33%), Positives = 70/133 (52%), Gaps = 7/133 (5%)
Query 8 NAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKDA 67
NA T +TV+ F V A + +++ FF +P RE AV++EF + LN D
Sbjct 102 NATTYGEATVYYFDVRPEAFEEAVDMFADFFKSPLLKRDSVEREVSAVNSEFCNGLNNDG 161
Query 68 LRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAVI 126
R ++++ K + S FS GN D+L R+ GI +E++ F + YSS+ M VI
Sbjct 162 WRTWRMMKKCCKKEQALSKFSTGNYDTL----RRDGI--WEEMKEFWSRKYSSDKMCVVI 215
Query 127 VGAESLDRLQEMV 139
G +S L+E++
Sbjct 216 YGNKSPGELKELL 228
> 7292708
Length=1077
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 42/148 (28%), Positives = 60/148 (40%), Gaps = 15/148 (10%)
Query 6 LANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNK 65
ANA T T+F F V L +L+ P + RE AV +EF L
Sbjct 145 FANANTDCEDTLFYFEVAEKHLDSSLDYFTALMKAPLMKQEAMQRERSAVDSEFQQILQD 204
Query 66 DALRLSYLIRQSKLKT-PFSSFSVGNLDSLVREPRKKGID------FLKEVRRFHDKWYS 118
D R L+ K P +F+ GN+ SL K+ +D L E+R+ H Y
Sbjct 205 DETRRDQLLASLATKGFPHGTFAWGNMKSL-----KENVDDAELHKILHEIRKEH---YG 256
Query 119 SNLMTAVIVGAESLDRLQEMVVNTLGKI 146
+N M + +D L+ +VV I
Sbjct 257 ANRMYVCLQARLPIDELESLVVRHFSGI 284
> 7296373
Length=908
Score = 43.5 bits (101), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 33/124 (26%), Positives = 54/124 (43%), Gaps = 9/124 (7%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLHKLNKD 66
+NA+T T F F + L + ++L P +RE AV +EF +D
Sbjct 146 SNAHTENEETCFYFELDQTHLDRGMDLFMNLMKAPLMLPDAMSRERSAVQSEFEQTHMRD 205
Query 67 ALRLSYLIRQ-SKLKTPFSSFSVGNLDSLVREPRKKGID---FLKEVRRFHDKWYSSNLM 122
+R ++ + P +FS GN +L K+G+D KE+ +F+ Y SN M
Sbjct 206 EVRRDQILASLASEGYPHGTFSWGNYKTL-----KEGVDDSSLHKELHKFYRDHYGSNRM 260
Query 123 TAVI 126
+
Sbjct 261 VVAL 264
> YPR122w
Length=1208
Score = 41.6 bits (96), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 35/146 (23%), Positives = 62/146 (42%), Gaps = 7/146 (4%)
Query 8 NAYTRRFSTVFSFAV------GAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHAEFLH 61
NA+T T F F + G L++ FF P F+ ++E A+ +E
Sbjct 100 NAFTTGEQTTFYFELPNTQNNGEFTFESILDVFASFFKEPLFNPLLISKEIYAIQSEHEG 159
Query 62 KLNKDALRLSYLIR-QSKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDKWYSSN 120
++ + R + PFS FS GN+ SL P+ K I + + + +
Sbjct 160 NISSTTKIFYHAARILANPDHPFSRFSTGNIHSLSSIPQLKKIKLKSSLNTYFENNFFGE 219
Query 121 LMTAVIVGAESLDRLQEMVVNTLGKI 146
+T I G +S++ L ++ ++ G I
Sbjct 220 NITLCIRGPQSVNILTKLALSKFGDI 245
> At4g00190
Length=474
Score = 33.1 bits (74), Expect = 0.20, Method: Composition-based stats.
Identities = 17/67 (25%), Positives = 38/67 (56%), Gaps = 1/67 (1%)
Query 37 FFLNPRFSAKPSNRETLAVHAEFLHKLNKDALRLSYLIRQSKLKTPFSSFSVGNLDSLVR 96
+FL P+F +K + E + ++ + +H+LN+ L + +S ++ S+ ++ NLD+
Sbjct 53 YFLEPQFGSKQAWEECMDLYEQTIHRLNESVLCPKNVCSRSDVQAWLST-ALTNLDTCQE 111
Query 97 EPRKKGI 103
E + G+
Sbjct 112 EMSELGV 118
> At5g42390
Length=1265
Score = 33.1 bits (74), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 67/149 (44%), Gaps = 29/149 (19%)
Query 7 ANAYTRRFSTVFSFAVGAPALSQ---------TLNLLKPFFLNPRFSAKPSNRETLAVHA 57
+NAYT TVF + +P ++ L+ L +P+F + +E A+ +
Sbjct 263 SNAYTDFHHTVFH--IHSPTHTKDSEDDLFPSVLDALNEIAFHPKFLSSRVEKERRAILS 320
Query 58 EFLHKLNK-----DALRLSYLIRQSKLKTPFSSFSVGNLDSLVREPRKKGIDFLKEVRRF 112
E L +N D L +L ++KL F +G L + +K +D ++R+F
Sbjct 321 E-LQMMNTIEYRVDCQLLQHLHSENKLG---RRFPIG----LEEQIKKWDVD---KIRKF 369
Query 113 HDKWYSSNLMTAVIVGAESLDRLQEMVVN 141
H++WY T IVG +D + +V N
Sbjct 370 HERWYFPANATLYIVG--DIDNIPRIVHN 396
> Hs22047356
Length=293
Score = 31.6 bits (70), Expect = 0.53, Method: Compositional matrix adjust.
Identities = 22/68 (32%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query 79 LKTPFSSFSV---GNLDSLVREPRKKGIDFLKEVRRFHDKWYSSNLMTAVIVGAESLDRL 135
L FS FS+ G S +R+P K +++L + DK+YS++L T + + S D
Sbjct 109 LTCTFSGFSLSTSGMCVSWIRQPPGKALEWLALIDWDDDKYYSTSLKTRLTI---SKDTS 165
Query 136 QEMVVNTL 143
+ VV T+
Sbjct 166 KNQVVLTM 173
> Hs4504931
Length=505
Score = 30.8 bits (68), Expect = 0.87, Method: Composition-based stats.
Identities = 27/109 (24%), Positives = 49/109 (44%), Gaps = 5/109 (4%)
Query 42 RFSAKPSNRETLAVHAEFLHKLNKDALR-LSYLIRQSKLKTPFSSFSVGNLDSLVREPRK 100
R S +N E L +FL +L ++ +R L I + + + N+D ++ E +
Sbjct 221 RKSDLEANVEALIQEIDFLRRLYEEEIRILQSHISDTSVVVKLDNSRDLNMDCIIAEIKA 280
Query 101 KGIDFLKEVRRFHDKWYSSN---LMTAVIVGAESLDRLQEMVVNTLGKI 146
+ D + R + WY S + VI E+L R +E +N L ++
Sbjct 281 QYDDIVTRSRAEAESWYRSKCEEMKATVIRHGETLRRTKEE-INELNRM 328
> Hs14318422
Length=486
Score = 30.8 bits (68), Expect = 0.87, Method: Composition-based stats.
Identities = 27/109 (24%), Positives = 49/109 (44%), Gaps = 5/109 (4%)
Query 42 RFSAKPSNRETLAVHAEFLHKLNKDALR-LSYLIRQSKLKTPFSSFSVGNLDSLVREPRK 100
R S +N E L +FL +L ++ +R L I + + + N+D ++ E +
Sbjct 221 RKSDLEANVEALIQEIDFLRRLYEEEIRVLQSHISDTSVVVKLDNSRDLNMDCIIAEIKA 280
Query 101 KGIDFLKEVRRFHDKWYSSN---LMTAVIVGAESLDRLQEMVVNTLGKI 146
+ D + R + WY S + VI E+L R +E +N L ++
Sbjct 281 QYDDIVTRSRAEAESWYRSKCEEMKATVIRHGETLRRTKEE-INELNRM 328
> Hs15431323
Length=493
Score = 30.4 bits (67), Expect = 1.2, Method: Composition-based stats.
Identities = 28/109 (25%), Positives = 48/109 (44%), Gaps = 5/109 (4%)
Query 42 RFSAKPSNRETLAVHAEFLHKLNKDALR-LSYLIRQSKLKTPFSSFSVGNLDSLVREPRK 100
R S +N E L +FL +L ++ +R L I + + + N+D +V E +
Sbjct 226 RKSDLEANVEALIQEIDFLRRLYEEEIRILQSHISDTSVVVKLDNSRDLNMDCIVAEIKA 285
Query 101 KGIDFLKEVRRFHDKWYSSN---LMTAVIVGAESLDRLQEMVVNTLGKI 146
+ D R + WY S + VI E+L R +E +N L ++
Sbjct 286 QYDDIATRSRAEAESWYRSKCEEMKATVIRHGETLRRTKEE-INELNRM 333
> 7297089
Length=506
Score = 29.3 bits (64), Expect = 2.7, Method: Composition-based stats.
Identities = 27/106 (25%), Positives = 49/106 (46%), Gaps = 11/106 (10%)
Query 16 TVFSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLAVHA----EFLHKLNKDALRLS 71
T+F G L++ KP P F +PSN + + +H + + L +D+
Sbjct 367 TLFITHAGKGGLTEAQYHGKPMLALPVFGDQPSNADVMVMHGFGIKQSILTLEEDSFLQG 426
Query 72 YLIRQSKLKTPFSSFSVGNLDSLVRE----PRKKGIDFLKEVRRFH 113
IR+ L P + +V + +L R+ PR+ I +++ V R+H
Sbjct 427 --IREV-LDNPKYATAVKSFSTLYRDRPLSPRETLIYWVEYVIRYH 469
> At1g16630
Length=845
Score = 28.1 bits (61), Expect = 6.2, Method: Composition-based stats.
Identities = 26/94 (27%), Positives = 46/94 (48%), Gaps = 6/94 (6%)
Query 28 SQTLNLLKPFFLNPRFSAKPSNRETL----AVHAEFLHKLNK-DALRLSYLIR-QSKLKT 81
S+T +L++ F FSA PS R ++ + E K N+ D +++ +++ +K
Sbjct 36 SETGDLMRRSFRGNPFSADPSRRNSIGRECSNRVEIGDKENQNDKDQIANVVKGPTKGSK 95
Query 82 PFSSFSVGNLDSLVREPRKKGIDFLKEVRRFHDK 115
F S ++ + + PRKK + EV R DK
Sbjct 96 HFMSPTISAVSKINPSPRKKILSDKNEVSRSFDK 129
> SPAC19B12.03
Length=1826
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Query 18 FSFAVGAPALSQTLNLLKPFFLNPRFSAKPSNRETLA 54
F F++G A + L PF L PRF + S+R+ LA
Sbjct 543 FFFSIGCVAYQSFIPL--PFLLGPRFKFRSSSRKYLA 577
> CE09454
Length=726
Score = 27.7 bits (60), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 22/86 (25%), Positives = 38/86 (44%), Gaps = 8/86 (9%)
Query 53 LAVHAEFLHKLNKDALRLSYLIRQSKLKTPFSSFSVGNL---DSLVREPRKKGIDFLKEV 109
L+VH + K N + L + YL + S + F GNL D + E +K I+ L+E
Sbjct 380 LSVHDNVIKKRNCEKLVIEYLPKASLRVFVRNHFEKGNLKIVDEMAEETKKSFIEMLQE- 438
Query 110 RRFHDKWYSSNLMTAVIVGAESLDRL 135
W ++ I+ E + ++
Sbjct 439 ----STWLGADSKKNAILKLEKMGKM 460
Lambda K H
0.323 0.136 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1748847648
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40