bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0636_orf2
Length=114
Score E
Sequences producing significant alignments: (Bits) Value
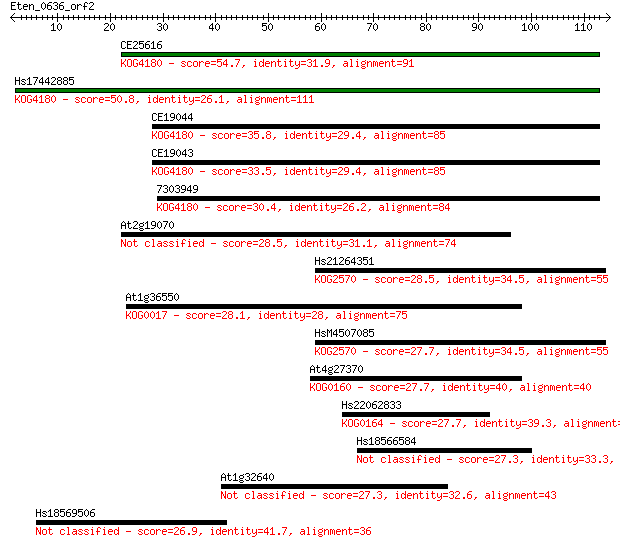
CE25616 54.7 4e-08
Hs17442885 50.8 7e-07
CE19044 35.8 0.022
CE19043 33.5 0.097
7303949 30.4 0.92
At2g19070 28.5 3.3
Hs21264351 28.5 3.3
At1g36550 28.1 3.9
HsM4507085 27.7 4.8
At4g27370 27.7 5.0
Hs22062833 27.7 5.3
Hs18566584 27.3 6.2
At1g32640 27.3 7.1
Hs18569506 26.9 8.4
> CE25616
Length=436
Score = 54.7 bits (130), Expect = 4e-08, Method: Composition-based stats.
Identities = 29/91 (31%), Positives = 47/91 (51%), Gaps = 0/91 (0%)
Query 22 PLSDAELQKVVQKANEQLLLDPAAPLMRFLVREPIDNRIFSCERTMGTGRNIRIKPLAGE 81
P++ ++KVV NE++ +P P F +REPI N + T G R I+++
Sbjct 343 PITRNLVEKVVNSVNEKIPFEPDHPSFAFSIREPIFNATYKRTATRGFARKIKLESRCTN 402
Query 82 IILSMDGLAELPLPPLVEIALSVDPSDALWT 112
L +DG ++P P + V+P+DAL T
Sbjct 403 GYLVLDGSTKIPFPRGAVATIEVNPNDALKT 433
> Hs17442885
Length=279
Score = 50.8 bits (120), Expect = 7e-07, Method: Compositional matrix adjust.
Identities = 29/111 (26%), Positives = 52/111 (46%), Gaps = 0/111 (0%)
Query 2 AKAIVDQVLSLRPKIKDMEGPLSDAELQKVVQKANEQLLLDPAAPLMRFLVREPIDNRIF 61
A V+ VL++ + ++ PL+ ++KV + NE LL P P + F +REPI NR+F
Sbjct 164 ATQAVEDVLNIAKRQGNLSLPLNRELVEKVTNEYNESLLYSPEEPKILFSIREPIANRVF 223
Query 62 SCERTMGTGRNIRIKPLAGEIILSMDGLAELPLPPLVEIALSVDPSDALWT 112
S R + ++ + + +DG ++ ++ D L T
Sbjct 224 SSSRQRCFSSKVCVRSRCWDACMVVDGGTSFEFNDGAIASMMINKEDELRT 274
> CE19044
Length=492
Score = 35.8 bits (81), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 25/85 (29%), Positives = 35/85 (41%), Gaps = 0/85 (0%)
Query 28 LQKVVQKANEQLLLDPAAPLMRFLVREPIDNRIFSCERTMGTGRNIRIKPLAGEIILSMD 87
+ ++ K N+QL+ DP + F VR+PI N F G I IK + L +D
Sbjct 404 VSEICTKFNQQLIFDPDRKQLAFSVRDPIFNATFPPTDPRGFADKICIKSRGYDAHLVID 463
Query 88 GLAELPLPPLVEIALSVDPSDALWT 112
G E + V D L T
Sbjct 464 GGISYRFNDGSEAVMEVRDEDTLRT 488
> CE19043
Length=514
Score = 33.5 bits (75), Expect = 0.097, Method: Composition-based stats.
Identities = 25/85 (29%), Positives = 35/85 (41%), Gaps = 0/85 (0%)
Query 28 LQKVVQKANEQLLLDPAAPLMRFLVREPIDNRIFSCERTMGTGRNIRIKPLAGEIILSMD 87
+ ++ K N+QL+ DP + F VR+PI N F G I IK + L +D
Sbjct 426 VSEICTKFNQQLIFDPDRKQLAFSVRDPIFNATFPPTDPRGFADKICIKSRGYDAHLVID 485
Query 88 GLAELPLPPLVEIALSVDPSDALWT 112
G E + V D L T
Sbjct 486 GGISYRFNDGSEAVMEVRDEDTLRT 510
> 7303949
Length=409
Score = 30.4 bits (67), Expect = 0.92, Method: Composition-based stats.
Identities = 22/87 (25%), Positives = 39/87 (44%), Gaps = 3/87 (3%)
Query 29 QKVVQKANEQLLLDPAAPLMRFLVREPIDNRIFSCERTMGTG---RNIRIKPLAGEIILS 85
+++ Q+ N+ LL P P + + +RE I ++ +T + + +K + L
Sbjct 319 EEIAQRYNQGLLFAPDDPRLCYSIREQICVGVWPSPKTFKERDFVQTVFVKSHCIDANLV 378
Query 86 MDGLAELPLPPLVEIALSVDPSDALWT 112
+DG P + L V P DAL T
Sbjct 379 IDGSISFPFNDGAKALLEVHPEDALLT 405
> At2g19070
Length=451
Score = 28.5 bits (62), Expect = 3.3, Method: Composition-based stats.
Identities = 23/75 (30%), Positives = 33/75 (44%), Gaps = 7/75 (9%)
Query 22 PLSDAELQKVVQKANEQLLLDPAAPLMRFLVREPIDNRIFSCE-RTMGTGRNIRIKPLAG 80
PLS ++LQK+ KAN DPA R+ E + ++ C + G +P A
Sbjct 236 PLSTSQLQKLRSKANGSKHSDPAKGFTRY---ETVTGHVWRCACKARGHSPE---QPTAL 289
Query 81 EIILSMDGLAELPLP 95
I + E PLP
Sbjct 290 GICIDTRSRMEPPLP 304
> Hs21264351
Length=475
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 11/55 (20%)
Query 59 RIFSCERTMGTGRNIRIKPLAGEIILSMDGLAELPLPPLVEIALSVDPSDALWTA 113
+IFSC R +R EI + + GL + P P ++ +SVDP+D TA
Sbjct 305 QIFSCGR-------LRF----SEIPMKLAGLLQHPDPIVINHVISVDPNDQKKTA 348
> At1g36550
Length=353
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 21/75 (28%), Positives = 36/75 (48%), Gaps = 4/75 (5%)
Query 23 LSDAELQKVVQKANEQLLLDPAAPLMRFLVREPIDNRIFSCERTMGTGRNIRIKPLAGEI 82
+ D +L QK +E+L L P + V EP++ R + ++ G G + + + G
Sbjct 25 MMDTKLNAFRQKFHEKLELGQTRPGEQMRVNEPLEQRDEATDQYYGIGA-LHLTIVKGGT 83
Query 83 ILSMDGLAELPLPPL 97
I D L +L +PP
Sbjct 84 I---DVLEKLKIPPF 95
> HsM4507085
Length=475
Score = 27.7 bits (60), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 19/55 (34%), Positives = 28/55 (50%), Gaps = 11/55 (20%)
Query 59 RIFSCERTMGTGRNIRIKPLAGEIILSMDGLAELPLPPLVEIALSVDPSDALWTA 113
+IFSC R +R EI + + GL + P P ++ +SVDP+D TA
Sbjct 305 QIFSCGR-------LRF----SEIPMKLAGLLQHPDPIVINHVISVDPNDQKKTA 348
> At4g27370
Length=1126
Score = 27.7 bits (60), Expect = 5.0, Method: Composition-based stats.
Identities = 16/40 (40%), Positives = 20/40 (50%), Gaps = 4/40 (10%)
Query 58 NRIFSCERTMGTGRNIRIKPLAGEIILSMDGLAELPLPPL 97
N F ER GR RIK AGE++ + +G E PL
Sbjct 618 NSCFKGER----GRGFRIKHYAGEVLYNTNGFLEKNRDPL 653
> Hs22062833
Length=930
Score = 27.7 bits (60), Expect = 5.3, Method: Composition-based stats.
Identities = 11/28 (39%), Positives = 19/28 (67%), Gaps = 0/28 (0%)
Query 64 ERTMGTGRNIRIKPLAGEIILSMDGLAE 91
++TM GR+ RIK AG++ S++G +
Sbjct 418 DKTMEFGRDFRIKHYAGDVTYSVEGFID 445
> Hs18566584
Length=317
Score = 27.3 bits (59), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 11/33 (33%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 67 MGTGRNIRIKPLAGEIILSMDGLAELPLPPLVE 99
+G +R+ PL G++I + GL++LP P +
Sbjct 233 VGGALRLRVAPLQGKVICTPAGLSQLPTSPRFQ 265
> At1g32640
Length=623
Score = 27.3 bits (59), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 20/43 (46%), Gaps = 0/43 (0%)
Query 41 LDPAAPLMRFLVREPIDNRIFSCERTMGTGRNIRIKPLAGEII 83
LDP + + P N FS E T + +KP +GEI+
Sbjct 325 LDPTPSPVHSQTQNPKFNNTFSRELNFSTSSSTLVKPRSGEIL 367
> Hs18569506
Length=379
Score = 26.9 bits (58), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 15/36 (41%), Positives = 19/36 (52%), Gaps = 0/36 (0%)
Query 6 VDQVLSLRPKIKDMEGPLSDAELQKVVQKANEQLLL 41
VDQVL+ I+D GPL + +VV A LL
Sbjct 328 VDQVLNQNQNIRDRTGPLCRDRVTEVVNAAKVMKLL 363
Lambda K H
0.318 0.137 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1181971906
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40