bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
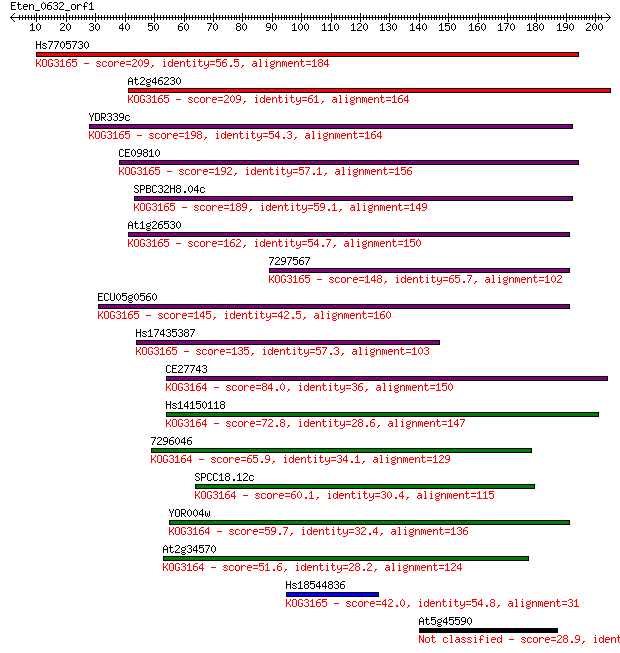
Query= Eten_0632_orf1
Length=204
Score E
Sequences producing significant alignments: (Bits) Value
Hs7705730 209 2e-54
At2g46230 209 4e-54
YDR339c 198 8e-51
CE09810 192 4e-49
SPBC32H8.04c 189 2e-48
At1g26530 162 6e-40
7297567 148 6e-36
ECU05g0560 145 7e-35
Hs17435387 135 6e-32
CE27743 84.0 2e-16
Hs14150118 72.8 4e-13
7296046 65.9 5e-11
SPCC18.12c 60.1 3e-09
YOR004w 59.7 4e-09
At2g34570 51.6 1e-06
Hs18544836 42.0 0.001
At5g45590 28.9 8.3
> Hs7705730
Length=198
Score = 209 bits (533), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 104/189 (55%), Positives = 138/189 (73%), Gaps = 5/189 (2%)
Query 10 TRRYMSTAQ---LRISCFRQKKEAKKQEKRRESETALKEH--VQHNPSQFFSYNTNLVPP 64
TR+Y + + LR ++K K ++K ++ +ALKE QH FF YNT L PP
Sbjct 7 TRKYATMKRMLSLRDQRLKEKDRLKPKKKEKKDPSALKEREVPQHPSCLFFQYNTQLGPP 66
Query 65 YQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDCVVAELEKLGHRYRLALHLAKD 124
Y +LVDTNFIN +I+ K D++Q ++ CL AKC C+TDCV+AE+EKLG +YR+AL +AKD
Sbjct 67 YHILVDTNFINFSIKAKLDLVQSMMDCLYAKCIPCITDCVMAEIEKLGQKYRVALRIAKD 126
Query 125 PRIKRLKCLHAGTYADDCIVQRVTEHKCYLVATNDKDLKRRIRKIPGVPIVYLKGRRFAV 184
PR +RL C H GTYADDC+VQRVT+HKCY+VAT D+DLKRRIRKIPGVPI+Y+ R+ +
Sbjct 127 PRFERLPCTHKGTYADDCLVQRVTQHKCYIVATVDRDLKRRIRKIPGVPIMYISNHRYNI 186
Query 185 ERLPEAITA 193
ER+P+ A
Sbjct 187 ERMPDDYGA 195
> At2g46230
Length=216
Score = 209 bits (531), Expect = 4e-54, Method: Compositional matrix adjust.
Identities = 100/169 (59%), Positives = 128/169 (75%), Gaps = 5/169 (2%)
Query 41 TALKEHVQHNPSQ-FFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVC 99
T L +V P+ FFSYN+ LVPPY+VLVDTNFIN +IQ K D+ +G+ CL A C C
Sbjct 38 TELPRNVPSVPAGLFFSYNSTLVPPYRVLVDTNFINFSIQNKIDLEKGMRDCLYANCTPC 97
Query 100 VTDCVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVTEHKCYLVATND 159
+TDCV+AELEKLG +YR+AL +AKDP +RL C+H GTYADDC+V RVT+HKC++VAT D
Sbjct 98 ITDCVMAELEKLGQKYRVALRIAKDPHFERLPCIHKGTYADDCLVDRVTQHKCFIVATCD 157
Query 160 KDLKRRIRKIPGVPIVYLKGRRFAVERLPEAITA----LPASKSGISKT 204
+DLKRRIRKIPGVPI+Y+ R++++E+LPEA LPA S + T
Sbjct 158 RDLKRRIRKIPGVPIMYVTNRKYSIEKLPEATLGGGMYLPAETSESTAT 206
> YDR339c
Length=189
Score = 198 bits (503), Expect = 8e-51, Method: Compositional matrix adjust.
Identities = 89/164 (54%), Positives = 123/164 (75%), Gaps = 0/164 (0%)
Query 28 KEAKKQEKRRESETALKEHVQHNPSQFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQG 87
K+ ++ K +E + Q + + FF YN + PPYQVL+DTNFIN +IQ K D+++G
Sbjct 26 KKNQENIKTKEDPELTRNIPQVSSALFFQYNQAIKPPYQVLIDTNFINFSIQKKVDIVRG 85
Query 88 LITCLVAKCDVCVTDCVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRV 147
++ CL+AKC+ +TDCV+AELEKLG +YR+AL LA+DPRIKRL C H GTYADDC+V RV
Sbjct 86 MMDCLLAKCNPLITDCVMAELEKLGPKYRIALKLARDPRIKRLSCSHKGTYADDCLVHRV 145
Query 148 TEHKCYLVATNDKDLKRRIRKIPGVPIVYLKGRRFAVERLPEAI 191
+HKCY+VATND LK+RIRKIPG+P++ + G + +E+LP+
Sbjct 146 LQHKCYIVATNDAGLKQRIRKIPGIPLMSVGGHAYVIEKLPDVF 189
> CE09810
Length=196
Score = 192 bits (488), Expect = 4e-49, Method: Compositional matrix adjust.
Identities = 89/156 (57%), Positives = 118/156 (75%), Gaps = 1/156 (0%)
Query 38 ESETALKEHVQHNPSQFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCD 97
E E L + + + F YNT L PP+ V+VDTNF+N A++ + D+ QG + CL AK
Sbjct 39 EQELKLVRAPKISSAMFMKYNTQLGPPFHVIVDTNFVNFAVKNRIDMFQGFMDCLFAKTI 98
Query 98 VCVTDCVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVTEHKCYLVAT 157
V TDCV+AELEK+ R+++AL + KDPR++RLKC H GTYADDC+VQRVT+HKCY+VAT
Sbjct 99 VYATDCVLAELEKV-RRFKIALKVLKDPRVQRLKCEHKGTYADDCLVQRVTQHKCYIVAT 157
Query 158 NDKDLKRRIRKIPGVPIVYLKGRRFAVERLPEAITA 193
D+DLKRRIRKIPGVPI+Y+ RF++ER+P+A A
Sbjct 158 CDRDLKRRIRKIPGVPIMYIVNHRFSIERMPDAYGA 193
> SPBC32H8.04c
Length=192
Score = 189 bits (481), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 88/149 (59%), Positives = 114/149 (76%), Gaps = 0/149 (0%)
Query 43 LKEHVQHNPSQFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTD 102
++E Q + FF +N +L PPY V++DTNFIN +Q K D+ +GL+TCL AK C++D
Sbjct 43 VREIPQMASNLFFQFNESLGPPYHVIIDTNFINFCLQQKIDLFEGLMTCLYAKTIPCISD 102
Query 103 CVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVTEHKCYLVATNDKDL 162
CV+AELEKLG RYR+AL +AKD R +RL C H GTYADDCIVQRV +HKCYLVATNDK+L
Sbjct 103 CVMAELEKLGIRYRIALRIAKDERFERLPCTHKGTYADDCIVQRVMQHKCYLVATNDKNL 162
Query 163 KRRIRKIPGVPIVYLKGRRFAVERLPEAI 191
K+RIRKIPG+PI+ + + VERL + +
Sbjct 163 KQRIRKIPGIPILSVANHKIRVERLVDVV 191
> At1g26530
Length=189
Score = 162 bits (409), Expect = 6e-40, Method: Compositional matrix adjust.
Identities = 82/151 (54%), Positives = 105/151 (69%), Gaps = 27/151 (17%)
Query 41 TALKEHVQHNPSQ-FFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVC 99
T L +V PS FFS+N++LVPPY+VLVDTNFIN +IQ K
Sbjct 38 TELPRNVPSVPSGLFFSHNSSLVPPYRVLVDTNFINFSIQNK------------------ 79
Query 100 VTDCVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVTEHKCYLVATND 159
LEKLG +YR+AL +AKDPR +RL C+ GTYADDC+V RVT+HKC++VAT D
Sbjct 80 --------LEKLGQKYRVALRIAKDPRFERLPCVLKGTYADDCLVDRVTQHKCFIVATCD 131
Query 160 KDLKRRIRKIPGVPIVYLKGRRFAVERLPEA 190
+DLKRRIRKIPGVPI+Y+ R++++E+LPEA
Sbjct 132 RDLKRRIRKIPGVPIMYVTQRKYSIEKLPEA 162
> 7297567
Length=107
Score = 148 bits (374), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 67/102 (65%), Positives = 85/102 (83%), Gaps = 0/102 (0%)
Query 89 ITCLVAKCDVCVTDCVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVT 148
+ CL AKC ++DCV AELEKLG++Y+LAL + DPR +RL CLH GTYADDC+V+RV
Sbjct 1 MDCLYAKCIPYISDCVRAELEKLGNKYKLALRIISDPRFERLPCLHKGTYADDCLVERVR 60
Query 149 EHKCYLVATNDKDLKRRIRKIPGVPIVYLKGRRFAVERLPEA 190
+HKCY+VATNDKDLK RIRKIPGVPI+Y+ ++A+ER+PEA
Sbjct 61 QHKCYIVATNDKDLKNRIRKIPGVPIMYVAAHKYAIERMPEA 102
> ECU05g0560
Length=179
Score = 145 bits (365), Expect = 7e-35, Method: Compositional matrix adjust.
Identities = 68/160 (42%), Positives = 102/160 (63%), Gaps = 0/160 (0%)
Query 31 KKQEKRRESETALKEHVQHNPSQFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLIT 90
+K+ + + E A ++ F N L PPY +++DTNFIN I+ K D+ + L+
Sbjct 18 RKKTRVKSPENASSTGENPRNTEDFLLNHTLRPPYNIILDTNFINDCIRKKYDIREQLMK 77
Query 91 CLVAKCDVCVTDCVVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVTEH 150
+ A +C+ DCV+ ELE LG +R+AL +D ++RL+C H GTYADDCI RV+ H
Sbjct 78 AVNASIKICIPDCVIGELESLGRPFRVALASIRDSDVQRLECDHKGTYADDCIFNRVSAH 137
Query 151 KCYLVATNDKDLKRRIRKIPGVPIVYLKGRRFAVERLPEA 190
+CY+VAT+D LK+RI+ IPGVP++ +G+R +ER A
Sbjct 138 RCYIVATSDVALKQRIKAIPGVPLITYRGQRCFIERFIPA 177
> Hs17435387
Length=136
Score = 135 bits (340), Expect = 6e-32, Method: Compositional matrix adjust.
Identities = 59/103 (57%), Positives = 77/103 (74%), Gaps = 0/103 (0%)
Query 44 KEHVQHNPSQFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDC 103
+E QH FF YNT L PPY +L+DTNFIN +I+ K D++Q ++ CL AKC C+TDC
Sbjct 34 REVPQHPSHLFFQYNTQLGPPYHILIDTNFINFSIKAKLDLMQSMMDCLYAKCIPCITDC 93
Query 104 VVAELEKLGHRYRLALHLAKDPRIKRLKCLHAGTYADDCIVQR 146
V+AE+EKLG +YR+AL + KDPR + L C H GTYADDC+VQ+
Sbjct 94 VMAEIEKLGQKYRVALRITKDPRFELLPCAHQGTYADDCLVQK 136
> CE27743
Length=232
Score = 84.0 bits (206), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 54/155 (34%), Positives = 82/155 (52%), Gaps = 13/155 (8%)
Query 54 FFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDCVVAELEKLGH 113
F+ YN V PY+VLVD F N A+Q K ++ + + L + + T+CV+ ELEK G
Sbjct 15 FYKYNYKFVAPYRVLVDGTFCNAALQEKLNLAEQIPKYLTEETHLMTTNCVLKELEKFGP 74
Query 114 RYRLALHLAKDPRIKRLKCLHAGT-YADDC---IVQRVTEHKC-YLVATNDKDLKRRIRK 168
A +AK I +C H+ A DC + +R K YL+AT D +L ++R
Sbjct 75 LLYGAFVIAKQFEIA--ECTHSTPRAASDCLAHLARRAASGKTKYLIATQDDELTEKLRA 132
Query 169 IPGVPIVYLKGRRFAVERLPEAITALPASKSGISK 203
I G PI+Y+K + ++ + E A+K+G SK
Sbjct 133 IVGTPIMYIKFKTVLLDNISE------ATKAGCSK 161
> Hs14150118
Length=249
Score = 72.8 bits (177), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 42/150 (28%), Positives = 77/150 (51%), Gaps = 3/150 (2%)
Query 54 FFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDCVVAELEKLGH 113
FF N + PYQ+L+D F A++ + + + L L+ + +C T CV+ ELE LG
Sbjct 15 FFRNNFGVREPYQILLDGTFCQAALRGRIQLREQLPRYLMGETQLCTTRCVLKELETLGK 74
Query 114 RYRLALHLAKDPRIKRLKCLHAGTYADDCIVQRVTEHKC--YLVATNDKDLKRRIRKIPG 171
A +A+ +++ +C++ V E Y VAT D++L +++K PG
Sbjct 75 DLYGAKLIAQKCQVRNCPHFKNAVSGSECLLSMVEEGNPHHYFVATQDQNLSVKVKKKPG 134
Query 172 VPIVYLKGRRFAVER-LPEAITALPASKSG 200
VP++++ +++ P+ I + A +SG
Sbjct 135 VPLMFIIQNTMVLDKPSPKTIAFVKAVESG 164
> 7296046
Length=244
Score = 65.9 bits (159), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 44/131 (33%), Positives = 65/131 (49%), Gaps = 5/131 (3%)
Query 49 HNPSQFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDCVVAEL 108
H FF+ N + PYQVL+D F A+Q K + + + + T CV+ E
Sbjct 10 HKTLVFFASNFDYREPYQVLIDATFCQAALQQKIGIDEQIKKYFQCGVKLLTTQCVILES 69
Query 109 EKLGHRYRLALHLAKDPRIKRLKCLHAG--TYADDCIVQRVTEHKCYLVATNDKDLKRRI 166
E LG A + K R KC H G A +CI + +T+ Y+VA+ D+ L+ +
Sbjct 70 ESLGAPLTGATSIVK--RFHVHKCGHEGKPVPASECI-KSMTKDNRYVVASQDRLLQESL 126
Query 167 RKIPGVPIVYL 177
RKIPG ++YL
Sbjct 127 RKIPGRCLLYL 137
> SPCC18.12c
Length=260
Score = 60.1 bits (144), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 59/120 (49%), Gaps = 6/120 (5%)
Query 64 PYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDCVVAELEKLGHRYRLALHLAK 123
PYQVLVD +F+ Q K D+ L + +T C + +L + + +AK
Sbjct 25 PYQVLVDADFLKDLSQQKIDIQAALARTVQGAIKPMITQCCIRQLYSKSDELKQEIRIAK 84
Query 124 DPRIKRLKCLHAGTYADDCIVQRVT-----EHKCYLVATNDKDLKRRIRKIPGVPIVYLK 178
+R + +CI V +H+ Y+VAT D +L++ +R +PGVP++Y+K
Sbjct 85 SFERRRCGHIDEALSPSECIQSVVNINGRNKHR-YVVATQDPELRQALRSVPGVPLIYMK 143
> YOR004w
Length=254
Score = 59.7 bits (143), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 44/146 (30%), Positives = 74/146 (50%), Gaps = 17/146 (11%)
Query 55 FSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAKCDVCVTDCVVAELEKLGHR 114
+S+ PYQVLVD + + ++ GL L A V +T C + L + R
Sbjct 16 YSHTFKFREPYQVLVDNQLVLECNNSNFNLPSGLKRTLQADVKVMITQCCIQALYET--R 73
Query 115 YRLALHLAKDPRIKRLKCLHAGTYAD-----DCI-----VQRVTEHKCYLVATNDKDLKR 164
A++LAK + +R +C H ++ D +CI + +H+ Y+VA+ D DL+R
Sbjct 74 NDGAINLAK--QFERRRCNH--SFKDPKSPAECIESVVNISGANKHR-YVVASQDIDLRR 128
Query 165 RIRKIPGVPIVYLKGRRFAVERLPEA 190
++R +PGVP+++L +E L A
Sbjct 129 KLRTVPGVPLIHLTRSVMVMEPLSTA 154
> At2g34570
Length=258
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 65/130 (50%), Gaps = 10/130 (7%)
Query 53 QFFSYNTNLVPPYQVLVDTNFINGAIQTKQDVLQGLITCLVAK-CDVCVTDCVVAELEKL 111
+FF+ PY+VL D F++ + + ++ L+ + T CV+AELEKL
Sbjct 14 RFFTVCYGFRQPYKVLCDGTFVHHLVTNEITPADTAVSELLGGPVKLFTTRCVIAELEKL 73
Query 112 GHRYRLALHLAKDPRIKRLKCLH-AGTYADDCIVQRV----TEHKCYLVATNDKDLKRRI 166
G + +L A+ + C H AD+C+ + + TEH + + T D + +R++
Sbjct 74 GKDFAESLEAAQT--LNTATCEHEEAKTADECLSEVIGVQNTEH--FFLGTQDAEFRRKL 129
Query 167 RKIPGVPIVY 176
++ VP+V+
Sbjct 130 QQESIVPLVF 139
> Hs18544836
Length=141
Score = 42.0 bits (97), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 17/31 (54%), Positives = 23/31 (74%), Gaps = 0/31 (0%)
Query 95 KCDVCVTDCVVAELEKLGHRYRLALHLAKDP 125
KC C+TDCV+AE+ KLG +Y A+ + KDP
Sbjct 108 KCIPCITDCVMAEIGKLGQKYGAAVRIVKDP 138
> At5g45590
Length=262
Score = 28.9 bits (63), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 17/47 (36%), Positives = 24/47 (51%), Gaps = 0/47 (0%)
Query 140 DDCIVQRVTEHKCYLVATNDKDLKRRIRKIPGVPIVYLKGRRFAVER 186
+D ++R E K + K KRR+R+ VP Y K + AVER
Sbjct 126 NDGTIRRWKEGKRHNAHLKSKKSKRRLRQPGLVPPAYAKDKTIAVER 172
Lambda K H
0.323 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3622842326
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40