bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0606_orf3
Length=201
Score E
Sequences producing significant alignments: (Bits) Value
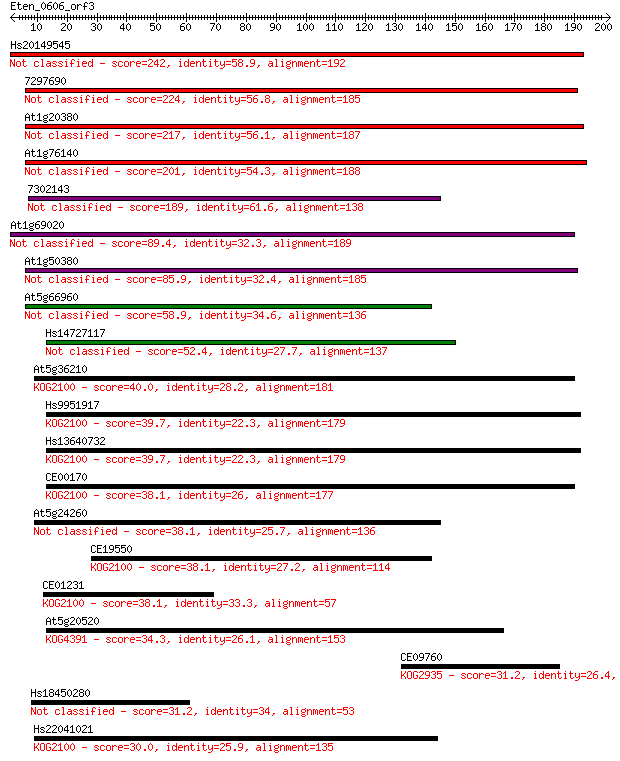
Hs20149545 242 5e-64
7297690 224 6e-59
At1g20380 217 1e-56
At1g76140 201 1e-51
7302143 189 4e-48
At1g69020 89.4 5e-18
At1g50380 85.9 4e-17
At5g66960 58.9 6e-09
Hs14727117 52.4 6e-07
At5g36210 40.0 0.003
Hs9951917 39.7 0.004
Hs13640732 39.7 0.004
CE00170 38.1 0.012
At5g24260 38.1 0.013
CE19550 38.1 0.013
CE01231 38.1 0.013
At5g20520 34.3 0.15
CE09760 31.2 1.3
Hs18450280 31.2 1.6
Hs22041021 30.0 3.1
> Hs20149545
Length=710
Score = 242 bits (617), Expect = 5e-64, Method: Compositional matrix adjust.
Identities = 113/192 (58%), Positives = 140/192 (72%), Gaps = 0/192 (0%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
I KQ +DDFQ AAEYL K+GY+SP +L I GGSNGGLLV ACANQRPDLF V+A
Sbjct 517 GGILANKQNCFDDFQCAAEYLIKEGYTSPKRLTINGGSNGGLLVAACANQRPDLFGCVIA 576
Query 61 KVGVMDLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVM 120
+VGVMD+L+FHK+TIGHAW +DYG D ++ F + K SPLHN+K A + P++L++
Sbjct 577 QVGVMDMLKFHKYTIGHAWTTDYGCSDSKQHFEWLVKYSPLHNVKLPEADDIQYPSMLLL 636
Query 121 TADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADT 180
TADHDDRV P HSLK+IA LQ+ G S S PL+ H+DTKAGHG GKP KV+EE +D
Sbjct 637 TADHDDRVVPLHSLKFIATLQYIVGRSRKQSNPLLIHVDTKAGHGAGKPTAKVIEEVSDM 696
Query 181 YGFIASVLGLKW 192
+ FIA L + W
Sbjct 697 FAFIARCLNVDW 708
> 7297690
Length=717
Score = 224 bits (572), Expect = 6e-59, Method: Compositional matrix adjust.
Identities = 105/185 (56%), Positives = 132/185 (71%), Gaps = 0/185 (0%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KQ +DDFQ+AAEYL + Y++ D+LAI GGSNGGLLVG+C NQRPDLF A VA+VGVM
Sbjct 528 NKQNVFDDFQAAAEYLIENKYTTKDRLAIQGGSNGGLLVGSCINQRPDLFGAAVAQVGVM 587
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHD 125
D+LRFHKFTIGHAW SDYG+P ++E F ++YK SPLHN+ + P+ L++TADHD
Sbjct 588 DMLRFHKFTIGHAWCSDYGNPSEKEHFDNLYKFSPLHNVHTPKGAETEYPSTLILTADHD 647
Query 126 DRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIA 185
DRVSP HSLK+IA LQ A S P++ + KAGHG GKP K +EE D F++
Sbjct 648 DRVSPLHSLKFIAALQEAVRDSEFQKNPVLLRVYQKAGHGAGKPTSKRIEEATDILTFLS 707
Query 186 SVLGL 190
L +
Sbjct 708 KSLNV 712
> At1g20380
Length=731
Score = 217 bits (552), Expect = 1e-56, Method: Compositional matrix adjust.
Identities = 105/194 (54%), Positives = 136/194 (70%), Gaps = 7/194 (3%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KQ +DDF S AEYL GY+ P +L I GGSNGG+LVGAC NQRPDLF +A VGVM
Sbjct 536 NKQNCFDDFISGAEYLVSAGYTQPRKLCIEGGSNGGILVGACINQRPDLFGCALAHVGVM 595
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIK-PEVAQSD---KQPAVLVMT 121
D+LRFHKFTIGHAW S++G DKEE+F + K SPLHN+K P ++D + P+ +++T
Sbjct 596 DMLRFHKFTIGHAWTSEFGCSDKEEEFHWLIKYSPLHNVKRPWEQKTDLFFQYPSTMLLT 655
Query 122 ADHDDRVSPFHSLKYIAELQHAAGYS---SPVSIPLIAHIDTKAGHGGGKPLMKVLEEQA 178
ADHDDRV P HS K +A +Q+ G S SP + P+IA I+ KAGHG G+P K+++E A
Sbjct 656 ADHDDRVVPLHSYKLLATMQYELGLSLENSPQTNPIIARIEVKAGHGAGRPTQKMIDEAA 715
Query 179 DTYGFIASVLGLKW 192
D Y F+A ++ W
Sbjct 716 DRYSFMAKMVDASW 729
> At1g76140
Length=724
Score = 201 bits (510), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 102/195 (52%), Positives = 129/195 (66%), Gaps = 14/195 (7%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KKQ +DDF S AEYL GY+ P +L I GGSNGGLL RPDL+ +A VGVM
Sbjct 536 KKQNCFDDFISGAEYLVSAGYTQPSKLCIEGGSNGGLL-------RPDLYGCALAHVGVM 588
Query 66 DLLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIK-PEVAQSD---KQPAVLVMT 121
D+LRFHKFTIGHAW SDYG + EE+F + K SPLHN+K P Q+D + P+ +++T
Sbjct 589 DMLRFHKFTIGHAWTSDYGCSENEEEFHWLIKYSPLHNVKRPWEQQTDHLVQYPSTMLLT 648
Query 122 ADHDDRVSPFHSLKYIAELQHAAGYS---SPVSIPLIAHIDTKAGHGGGKPLMKVLEEQA 178
ADHDDRV P HSLK +A LQH S SP P+I I+ KAGHG G+P K+++E A
Sbjct 649 ADHDDRVVPLHSLKLLATLQHVLCTSLDNSPQMNPIIGRIEVKAGHGAGRPTQKMIDEAA 708
Query 179 DTYGFIASVLGLKWS 193
D Y F+A ++ W+
Sbjct 709 DRYSFMAKMVNASWT 723
> 7302143
Length=723
Score = 189 bits (479), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 85/138 (61%), Positives = 108/138 (78%), Gaps = 0/138 (0%)
Query 7 KQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMD 66
KQ ++DFQ+AAE+L K Y++ D+LAI G SNGGLLVGAC NQRPDLF A VA+VGVMD
Sbjct 584 KQNVFNDFQAAAEFLTKNNYTTKDRLAIQGASNGGLLVGACINQRPDLFGAAVAQVGVMD 643
Query 67 LLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDD 126
+LRFHKFTIGHAW SDYG+PD++ F ++ K SPLHN+ + + + P+ L++TADHDD
Sbjct 644 MLRFHKFTIGHAWCSDYGNPDEKVHFANLIKFSPLHNVHIPLNPNQEYPSTLILTADHDD 703
Query 127 RVSPFHSLKYIAELQHAA 144
RVSP HS K++A LQ A
Sbjct 704 RVSPLHSYKFVAALQEAV 721
> At1g69020
Length=798
Score = 89.4 bits (220), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 61/195 (31%), Positives = 94/195 (48%), Gaps = 15/195 (7%)
Query 1 AAIKEKKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
+ + KQ S DF +A+YL +KGY LA +G S G +L A N P LF+AV+
Sbjct 608 SGTRSLKQNSIQDFIYSAKYLVEKGYVHRHHLAAVGYSAGAILPAAAMNMHPSLFQAVIL 667
Query 61 KVGVMDLL------RFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQ 114
KV +D+L + H ++G+PD + DF I SP I+ +V
Sbjct 668 KVPFVDVLNTLSDPNLPLTLLDH---EEFGNPDNQTDFGSILSYSPYDKIRKDVC----Y 720
Query 115 PAVLVMTADHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVL 174
P++LV T+ HD RV + K++A+++ + + S +I + GH G
Sbjct 721 PSMLVTTSFHDSRVGVWEGAKWVAKIRDSTCHD--CSRAVILKTNMNGGHFGEGGRYAQC 778
Query 175 EEQADTYGFIASVLG 189
EE A Y F+ V+G
Sbjct 779 EETAFDYAFLLKVMG 793
> At1g50380
Length=710
Score = 85.9 bits (211), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 60/188 (31%), Positives = 98/188 (52%), Gaps = 12/188 (6%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KK+ ++ DF + AE L + Y S ++L + G S GGLL+GA N RPDLF+ V+A V +
Sbjct 527 KKKNTFTDFIACAERLIELKYCSKEKLCMEGRSAGGLLMGAVVNMRPDLFKVVIAGVPFV 586
Query 66 DLLRFH---KFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTA 122
D+L + + ++GDP KEE + ++ SP+ N+ AQ+ P +LV
Sbjct 587 DVLTTMLDPTIPLTTSEWEEWGDPRKEEFYFYMKSYSPVDNV---TAQN--YPNMLVTAG 641
Query 123 DHDDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYG 182
+D RV K++A+L+ ++ L+ + AGH + L+E A T+
Sbjct 642 LNDPRVMYSEPGKWVAKLREMKTDNN----VLLFKCELGAGHFSKSGRFEKLQEDAFTFA 697
Query 183 FIASVLGL 190
F+ VL +
Sbjct 698 FMMKVLDM 705
> At5g66960
Length=792
Score = 58.9 bits (141), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 47/139 (33%), Positives = 67/139 (48%), Gaps = 8/139 (5%)
Query 6 KKQVSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVM 65
KK S D+ A+YL + ++LA G S GGL+V + N PDLF+A V KV +
Sbjct 615 KKLNSIKDYIQCAKYLVENNIVEENKLAGWGYSAGGLVVASAINHCPDLFQAAVLKVPFL 674
Query 66 DLLRFHKFTIGHAWVSDY---GDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTA 122
D + I DY G P DF I + SP NI +V PAVLV T+
Sbjct 675 DPTHTLIYPILPLTAEDYEEFGYPGDINDFHAIREYSPYDNIPKDVL----YPAVLV-TS 729
Query 123 DHDDRVSPFHSLKYIAELQ 141
+ R + + K++A ++
Sbjct 730 SFNTRFGVWEAAKWVARVR 748
> Hs14727117
Length=638
Score = 52.4 bits (124), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 38/150 (25%), Positives = 68/150 (45%), Gaps = 18/150 (12%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRF-- 70
D ++ + L +G+S P + S GG+L GA N P+L RAV + +D+L
Sbjct 445 DLEACIKTLHGQGFSQPSLTTLTAFSAGGVLAGALCNSNPELVRAVTLEAPFLDVLNTMM 504
Query 71 -HKFTIGHAWVSDYGDPDKEEDF-THIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRV 128
+ + ++G+P +E +I + P NIKP+ P++ + ++D+RV
Sbjct 505 DTTLPLTLEELEEWGNPSSDEKHKNYIKRYCPYQNIKPQ-----HYPSIHITAYENDERV 559
Query 129 SPFHSLKYIAELQHA---------AGYSSP 149
+ Y +L+ A GY +P
Sbjct 560 PLKGIVSYTEKLKEAIAEHAKDTGEGYQTP 589
> At5g36210
Length=678
Score = 40.0 bits (92), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 51/184 (27%), Positives = 80/184 (43%), Gaps = 24/184 (13%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V DD A+YL G + +L I GGS GG A R D+F+A + GV DL
Sbjct 498 VDVDDCCGCAKYLVSSGKADVKRLCISGGSAGGYTTLASLAFR-DVFKAGASLYGVADLK 556
Query 69 RFHKFTIGHAWVSDYGDP--DKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDD 126
+ GH + S Y D E+DF Y+ SP++ + DK +++ +D
Sbjct 557 MLKE--EGHKFESRYIDNLVGDEKDF---YERSPINFV-------DKFSCPIILFQGLED 604
Query 127 R-VSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIA 185
+ V+P S K L+ + + L+ + + G + + LE+Q F A
Sbjct 605 KVVTPDQSRKIYEALKKKG-----LPVALVEYEGEQHGFRKAENIKYTLEQQM---VFFA 656
Query 186 SVLG 189
V+G
Sbjct 657 RVVG 660
> Hs9951917
Length=732
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 40/180 (22%), Positives = 79/180 (43%), Gaps = 15/180 (8%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D Q A E + ++ + +A+MGGS+GG + Q P+ +RA VA+ V+++
Sbjct 562 DVQFAVEQVLQEEHFDASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLG 621
Query 73 FTIGHAW-VSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPF 131
T W V + G P + + + + + P + +L+M D RV
Sbjct 622 STDIPDWCVVEAGFPFSSDCLPDLSVWAEMLDKSPIRYIPQVKTPLLLMLGQEDRRVPFK 681
Query 132 HSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIASVLGLK 191
++Y L+ ++P+ + K+ H + +E ++D+ F+ +VL L+
Sbjct 682 QGMEYYRALKTR-------NVPVRLLLYPKSTHA-----LSEVEVESDS--FMNAVLWLR 727
> Hs13640732
Length=732
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 40/180 (22%), Positives = 79/180 (43%), Gaps = 15/180 (8%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D Q A E + ++ + +A+MGGS+GG + Q P+ +RA VA+ V+++
Sbjct 562 DVQFAVEQVLQEEHFDASHVALMGGSHGGFISCHLIGQYPETYRACVARNPVINIASMLG 621
Query 73 FTIGHAW-VSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPF 131
T W V + G P + + + + + P + +L+M D RV
Sbjct 622 STDIPDWCVVEAGFPFSSDCLPDLSVWAEMLDKSPIRYIPQVKTPLLLMLGQEDRRVPFK 681
Query 132 HSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFIASVLGLK 191
++Y L+ ++P+ + K+ H + +E ++D+ F+ +VL L+
Sbjct 682 QGMEYYRALKTR-------NVPVRLLLYPKSTHA-----LSEVEVESDS--FMNAVLWLR 727
> CE00170
Length=761
Score = 38.1 bits (87), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 46/185 (24%), Positives = 72/185 (38%), Gaps = 23/185 (12%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHK 72
D A E+ KG ++ ++A+MGGS GG P F V VG +L+ +
Sbjct 512 DILDAVEFAVSKGIANRSEVAVMGGSYGGYETLVALTFTPQTFACGVDIVGPSNLISLVQ 571
Query 73 FTIGHAWVSDYGDPDK--------EEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADH 124
I W+ D K EE + SPL A +P ++++ +
Sbjct 572 -AIPPYWLGFRKDLIKMVGADISDEEGRQSLQSRSPLF-----FADRVTKP-IMIIQGAN 624
Query 125 DDRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFI 184
D RV S +++A L+ IP+ + GHG KP +E+ F+
Sbjct 625 DPRVKQAESDQFVAALEKK-------HIPVTYLLYPDEGHGVRKP-QNSMEQHGHIETFL 676
Query 185 ASVLG 189
LG
Sbjct 677 QQCLG 681
> At5g24260
Length=746
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 61/137 (44%), Gaps = 13/137 (9%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V +D + A++L ++G + PD + + G S GG L + P++F V+ V
Sbjct 586 VDAEDQVTGAKWLIEQGLAKPDHIGVYGWSYGGYLSATLLTRYPEIFNCAVSGAPVTSWD 645
Query 69 RFHKFTIGHAWVSDY-GDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDR 127
+ F + Y G P +EE + K S +H++ +DKQ +LV D+
Sbjct 646 GYDSF-----YTEKYMGLPTEEERY---LKSSVMHHVG---NLTDKQKLMLVHGM-IDEN 693
Query 128 VSPFHSLKYIAELQHAA 144
V H+ + + L A
Sbjct 694 VHFRHTARLVNALVEAG 710
> CE19550
Length=745
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 50/119 (42%), Gaps = 9/119 (7%)
Query 28 SPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLLRFHKFTIGHAW-----VSD 82
S D++ + GGS+GG LV Q P +++ VA V+++ H T W +
Sbjct 590 SRDKVVLFGGSHGGFLVSHLIGQYPGFYKSCVALNPVVNIATMHDITDIPEWCYFEGTGE 649
Query 83 YGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDRVSPFHSLKYIAELQ 141
Y D K T + + N P + L++ + D RV P H +I L+
Sbjct 650 YPDWTK---ITTTEQREKMFNSSPIAHVENATTPYLLLIGEKDLRVVP-HYRAFIRALK 704
> CE01231
Length=629
Score = 38.1 bits (87), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 0/57 (0%)
Query 12 DDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
DD + A+ L ++G +++ I G S+GG L+ +C ++ +A V+ GV DLL
Sbjct 449 DDMLNGAKALVEQGKVDAEKVLITGSSSGGYLILSCLISPKNIIKAAVSVYGVADLL 505
> At5g20520
Length=340
Score = 34.3 bits (77), Expect = 0.15, Method: Compositional matrix adjust.
Identities = 40/160 (25%), Positives = 62/160 (38%), Gaps = 17/160 (10%)
Query 13 DFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAK---VGVMD--- 66
D Q+A ++L + ++ + G S GG + PD A++ + ++D
Sbjct 161 DAQAALDHLSGRTDIDTSRIVVFGRSLGGAVGAVLTKNNPDKVSALILENTFTSILDMAG 220
Query 67 -LLRFHKFTIGHAWVSDYGDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHD 125
LL F K+ IG G K + SP I + KQP VL ++ D
Sbjct 221 VLLPFLKWFIG-------GSGTKSLKLLNFVVRSPWKTI--DAIAEIKQP-VLFLSGLQD 270
Query 126 DRVSPFHSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHG 165
+ V PFH A+ + V P H+DT G
Sbjct 271 EMVPPFHMKMLYAKAAARNPQCTFVEFPSGMHMDTWLSGG 310
> CE09760
Length=317
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/53 (26%), Positives = 23/53 (43%), Gaps = 0/53 (0%)
Query 132 HSLKYIAELQHAAGYSSPVSIPLIAHIDTKAGHGGGKPLMKVLEEQADTYGFI 184
+L+ EL A G S P IPL+ + G +P +++ D F+
Sbjct 251 ENLRKAIELSQAPGPSEPAEIPLLTRSRSSTPPGASEPFSNAEQQRRDRQKFL 303
> Hs18450280
Length=882
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/54 (33%), Positives = 26/54 (48%), Gaps = 1/54 (1%)
Query 8 QVSYDDFQSAAEYLFKK-GYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVA 60
Q+ DD +YL + + D++ I G S GG L QR D+FR +A
Sbjct 708 QIEIDDQVEGLQYLASRYDFIDLDRVGIHGWSYGGYLSLMALMQRSDIFRVAIA 761
> Hs22041021
Length=746
Score = 30.0 bits (66), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 52/136 (38%), Gaps = 13/136 (9%)
Query 9 VSYDDFQSAAEYLFKKGYSSPDQLAIMGGSNGGLLVGACANQRPDLFRAVVAKVGVMDLL 68
V D +A ++L K Y +L+I G GG + LF+ + DL
Sbjct 572 VEVKDQITAVKFLLKLPYIDSKRLSIFGKGYGGYIASMILKSDEKLFKCGSVVAPITDLK 631
Query 69 RFHKFTIGHAWVSDY-GDPDKEEDFTHIYKISPLHNIKPEVAQSDKQPAVLVMTADHDDR 127
+ A+ Y G P KEE S LHN+ K+ +L++ D +
Sbjct 632 LY-----ASAFSERYLGMPSKEESTYQ--AASVLHNV-----HGLKEENILIIHGTADTK 679
Query 128 VSPFHSLKYIAELQHA 143
V HS + I L A
Sbjct 680 VHFQHSAELIKHLIKA 695
Lambda K H
0.317 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3515233148
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40