bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0589_orf1
Length=121
Score E
Sequences producing significant alignments: (Bits) Value
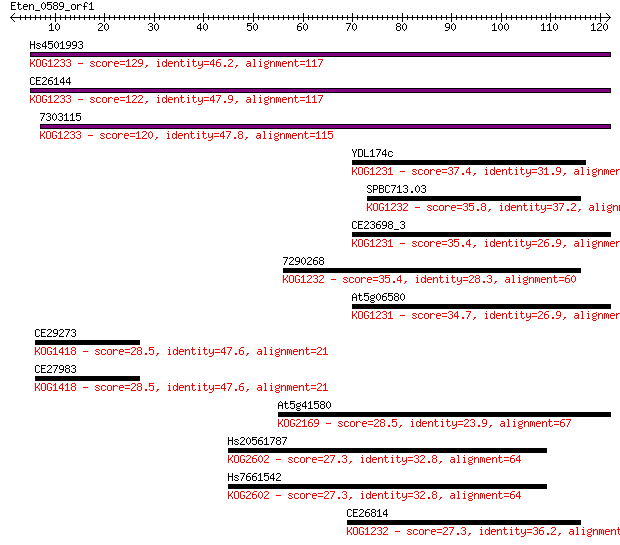
Hs4501993 129 2e-30
CE26144 122 2e-28
7303115 120 9e-28
YDL174c 37.4 0.007
SPBC713.03 35.8 0.018
CE23698_3 35.4 0.025
7290268 35.4 0.025
At5g06580 34.7 0.047
CE29273 28.5 3.1
CE27983 28.5 3.1
At5g41580 28.5 3.6
Hs20561787 27.3 6.8
Hs7661542 27.3 7.0
CE26814 27.3 7.6
> Hs4501993
Length=658
Score = 129 bits (323), Expect = 2e-30, Method: Compositional matrix adjust.
Identities = 54/117 (46%), Positives = 81/117 (69%), Gaps = 0/117 (0%)
Query 5 QRVKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN 64
+ VK+RI +C G+++ P R+TQ YD GA +YFYF FN+ G+ +P+ VF + E
Sbjct 542 RNVKERITRECKEKGVQFAPFSTCRVTQTYDAGACIYFYFAFNYRGISDPLTVFEQTEAA 601
Query 65 ARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
AR+ IL GS+SHHHGVGKLRK+++ +++ G +L++ K+ VDP NIFGN N++
Sbjct 602 AREEILANGGSLSHHHGVGKLRKQWLKESISDVGFGMLKSVKEYVDPNNIFGNRNLL 658
> CE26144
Length=597
Score = 122 bits (305), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 56/117 (47%), Positives = 78/117 (66%), Gaps = 0/117 (0%)
Query 5 QRVKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHN 64
+ VK+ +K + G+ +P L R+TQVYD GA +YFYFGFN GL+ ++V+ +E
Sbjct 471 RNVKELMKREAKAQGVTHPVLANCRVTQVYDAGACVYFYFGFNARGLKNGLEVYDRIETA 530
Query 65 ARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
AR I+ C GSISHHHGVGK+RK++M G G+ LL+A K +DP NIF + N+I
Sbjct 531 ARDEIIACGGSISHHHGVGKIRKQWMLTTNGAVGIALLKAIKSELDPANIFASANLI 587
> 7303115
Length=631
Score = 120 bits (300), Expect = 9e-28, Method: Compositional matrix adjust.
Identities = 55/115 (47%), Positives = 76/115 (66%), Gaps = 1/115 (0%)
Query 7 VKDRIKSDCSRLGIKYPPLVMVRLTQVYDGGAVLYFYFGFNWAGLREPIKVFSEVEHNAR 66
VK R+ S+CS+ I Y + R+TQ YD GA +YFYFGF + +P+++F +EH+AR
Sbjct 496 VKQRVVSECSKRSINYY-TISCRVTQTYDAGACIYFYFGFRSTDVADPVELFEAIEHSAR 554
Query 67 QVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
IL C GS+SHHHGVGK+R + AV +TG L A K +DP+NIF GN++
Sbjct 555 DEILSCGGSLSHHHGVGKIRSHWYRNAVTETGSSLYSAAKRHLDPKNIFALGNLL 609
> YDL174c
Length=587
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 15/47 (31%), Positives = 31/47 (65%), Gaps = 0/47 (0%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFG 116
L+ +G+ + HGVG ++E++ + +G+ ++L+R K A+DP+ I
Sbjct 524 LNAEGTCTGEHGVGIGKREYLLEELGEAPVDLMRKIKLAIDPKRIMN 570
> SPBC713.03
Length=526
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 25/43 (58%), Gaps = 0/43 (0%)
Query 73 KGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
+GSIS HG+G L+K F+ + K + L++ K+ DP I
Sbjct 478 RGSISAEHGLGLLKKPFVGYSKSKEMIHLMKTLKNVFDPNGIM 520
> CE23698_3
Length=454
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 30/52 (57%), Gaps = 0/52 (0%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
L G+ + HG+G +++++ + +G+ + L+ K A+DP NI G ++
Sbjct 400 LAADGTCTGEHGIGLGKRKYLREELGENTVRLMHTIKHALDPNNIMNPGKVL 451
> 7290268
Length=533
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 0/60 (0%)
Query 56 KVFSEVEHNARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIF 115
+++ VE + KGSIS HG+G L+K+++ + + +R K +DP +I
Sbjct 467 EIYKRVEPFVYEYTSKLKGSISAEHGIGFLKKDYLHYSKDPVAIGYMREMKKLLDPNSIL 526
> At5g06580
Length=417
Score = 34.7 bits (78), Expect = 0.047, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 28/52 (53%), Gaps = 0/52 (0%)
Query 70 LDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNIFGNGNMI 121
L G+ + HGVG + +++ K +G ++ ++ K +DP +I G +I
Sbjct 360 LSMDGTCTGEHGVGTGKMKYLEKELGIEALQTMKRIKKTLDPNDIMNPGKLI 411
> CE29273
Length=1702
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 10/21 (47%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 6 RVKDRIKSDCSRLGIKYPPLV 26
+V D+ K DCS+ GI PP++
Sbjct 432 KVMDKFKQDCSKKGIDPPPVL 452
> CE27983
Length=1526
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 10/21 (47%), Positives = 15/21 (71%), Gaps = 0/21 (0%)
Query 6 RVKDRIKSDCSRLGIKYPPLV 26
+V D+ K DCS+ GI PP++
Sbjct 432 KVMDKFKQDCSKKGIDPPPVL 452
> At5g41580
Length=719
Score = 28.5 bits (62), Expect = 3.6, Method: Composition-based stats.
Identities = 16/67 (23%), Positives = 30/67 (44%), Gaps = 12/67 (17%)
Query 55 IKVFSEVEHNARQVILDCKGSISHHHGVGKLRKEFMAKAVGKTGMELLRATKDAVDPQNI 114
+++ +VEHNA VI+D G+ + K G+T + D DP ++
Sbjct 330 LQILKDVEHNAADVIIDAGGTWK------------VTKNTGETPEPVREIIHDLEDPMSL 377
Query 115 FGNGNMI 121
+G ++
Sbjct 378 LNSGPVV 384
> Hs20561787
Length=469
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 9/72 (12%)
Query 45 GFNWAGLREPIKVFSEVEHNARQVILDCKGSISHH---HGVGKLRKEFMAKAVG-----K 96
G ++ GL E + F EVE A+Q IL+ K + H G+G+ + + + +G K
Sbjct 16 GPDFGGLGEEAE-FVEVEPEAKQEILENKDVVVQHVHFDGLGRTKDDIIICEIGDVFKAK 74
Query 97 TGMELLRATKDA 108
+E++R + +A
Sbjct 75 NLIEVMRKSHEA 86
> Hs7661542
Length=469
Score = 27.3 bits (59), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 9/72 (12%)
Query 45 GFNWAGLREPIKVFSEVEHNARQVILDCKGSISHH---HGVGKLRKEFMAKAVG-----K 96
G ++ GL E + F EVE A+Q IL+ K + H G+G+ + + + +G K
Sbjct 16 GPDFGGLGEEAE-FVEVEPEAKQEILENKDVVVQHVHFDGLGRTKDDIIICEIGDVFKAK 74
Query 97 TGMELLRATKDA 108
+E++R + +A
Sbjct 75 NLIEVMRKSHEA 86
> CE26814
Length=487
Score = 27.3 bits (59), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 17/49 (34%), Positives = 25/49 (51%), Gaps = 4/49 (8%)
Query 69 ILDCKGSISHHHGVGKLRKEFMAKAVGKTGME--LLRATKDAVDPQNIF 115
++D GSIS HG+G+L+ + GK E L + K+ DP I
Sbjct 435 VVDHGGSISAEHGIGQLKLPY--STFGKDPEERLLTKKLKNIFDPNGIL 481
Lambda K H
0.324 0.141 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1198419392
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40