bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0576_orf3
Length=268
Score E
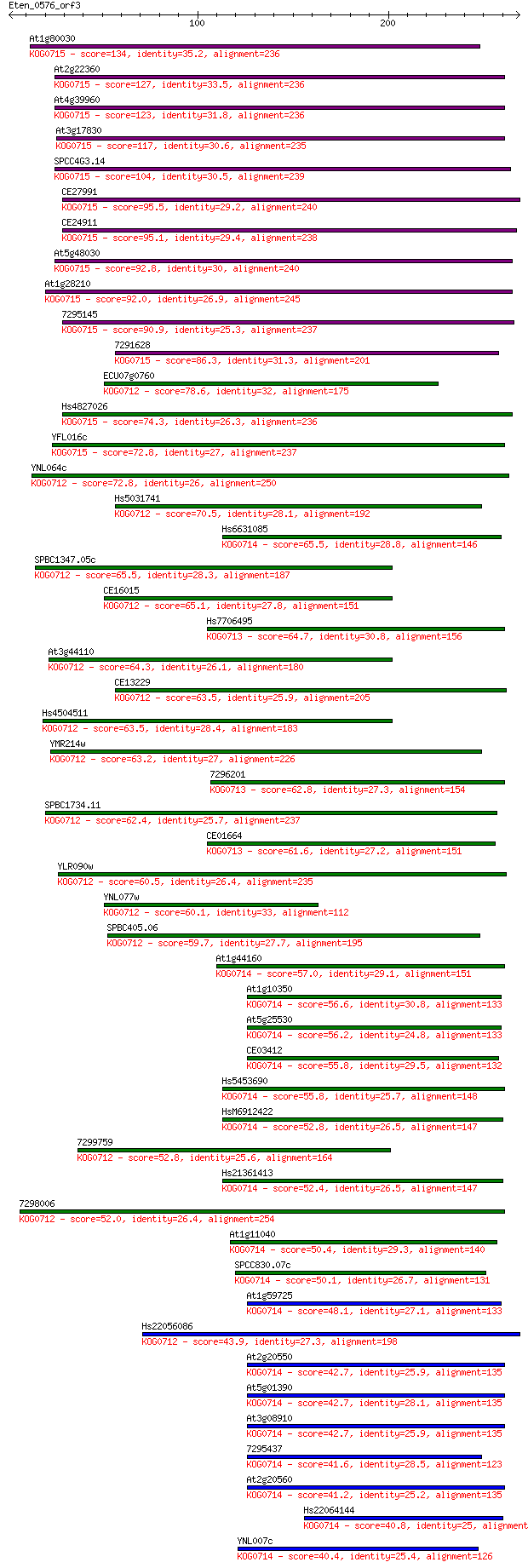
Sequences producing significant alignments: (Bits) Value
At1g80030 134 2e-31
At2g22360 127 2e-29
At4g39960 123 5e-28
At3g17830 117 3e-26
SPCC4G3.14 104 1e-22
CE27991 95.5 1e-19
CE24911 95.1 1e-19
At5g48030 92.8 6e-19
At1g28210 92.0 1e-18
7295145 90.9 2e-18
7291628 86.3 6e-17
ECU07g0760 78.6 1e-14
Hs4827026 74.3 2e-13
YFL016c 72.8 7e-13
YNL064c 72.8 7e-13
Hs5031741 70.5 3e-12
Hs6631085 65.5 1e-10
SPBC1347.05c 65.5 1e-10
CE16015 65.1 2e-10
Hs7706495 64.7 2e-10
At3g44110 64.3 3e-10
CE13229 63.5 4e-10
Hs4504511 63.5 4e-10
YMR214w 63.2 5e-10
7296201 62.8 7e-10
SPBC1734.11 62.4 9e-10
CE01664 61.6 2e-09
YLR090w 60.5 4e-09
YNL077w 60.1 5e-09
SPBC405.06 59.7 6e-09
At1g44160 57.0 4e-08
At1g10350 56.6 6e-08
At5g25530 56.2 6e-08
CE03412 55.8 9e-08
Hs5453690 55.8 1e-07
HsM6912422 52.8 8e-07
7299759 52.8 8e-07
Hs21361413 52.4 1e-06
7298006 52.0 1e-06
At1g11040 50.4 3e-06
SPCC830.07c 50.1 4e-06
At1g59725 48.1 2e-05
Hs22056086 43.9 4e-04
At2g20550 42.7 7e-04
At5g01390 42.7 7e-04
At3g08910 42.7 8e-04
7295437 41.6 0.002
At2g20560 41.2 0.003
Hs22064144 40.8 0.003
YNL007c 40.4 0.003
> At1g80030
Length=435
Score = 134 bits (337), Expect = 2e-31, Method: Compositional matrix adjust.
Identities = 83/239 (34%), Positives = 119/239 (49%), Gaps = 6/239 (2%)
Query 12 AAAGRRHTQGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTRRTSP- 70
A GR +G D+ +TL +A + ETC ACAG + S
Sbjct 179 ADFGRTRRSRVTKGEDLRYDITLELSEAIFGSEKEFDLT--HLETCEACAGTGAKAGSKM 236
Query 71 --CPVCEGQGMQTETRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKV 128
C C G+G T ++ FG + C C G C+ C GEG + +KV
Sbjct 237 RICSTCGGRGQVMRTEQTPFGMFSQVSICPNCGGDGEVISENCRKCSGEGRVRIKKSIKV 296
Query 129 NIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDA 188
IP GV GS+LRV GEG G G GDLY+ + VE + R+G ++ S SIS DA
Sbjct 297 KIPPGVSAGSILRVAGEGDSGPRGGPPGDLYVYLDVEDVRGIERDGINLLSTLSISYLDA 356
Query 189 VYGCSITVHTVDGDVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPE 247
+ G + V TV+GD EL+IP ++ G ++ +G K++ PS RGDH+ T++V++P
Sbjct 357 ILGAVVKVKTVEGDTELQIPPGTQPGDVLVLAKKGVPKLNRPS-IRGDHLFTVKVSVPN 414
> At2g22360
Length=391
Score = 127 bits (319), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 79/240 (32%), Positives = 127/240 (52%), Gaps = 9/240 (3%)
Query 25 GLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGMQ 80
G D + L+F +A + + R E+C C G P T+ T C C GQG
Sbjct 143 GQDEYYTLILNFKEAVF--GMEKEIEISRLESCGTCEGSGAKPGTKPTK-CTTCGGQGQV 199
Query 81 TETRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLL 140
R+ G +TCS+CN TG P C +C G+G + K + + +PAGV GS L
Sbjct 200 VSAARTPLGVFQQVMTCSSCNGTGEISTP-CGTCSGDGRVRKTKRISLKVPAGVDSGSRL 258
Query 141 RVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVD 200
RV GEG+ G+ G + GDL++ I V L R+ ++ IS DA+ G ++ V TVD
Sbjct 259 RVRGEGNAGKRGGSPGDLFVVIEVIPDPILKRDDTNILYTCKISYIDAILGTTLKVPTVD 318
Query 201 GDVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSL 260
G V+LK+PA ++ ++ +G ++ S RGD +V ++V +P+ + ++++L++ L
Sbjct 319 GTVDLKVPAGTQPSTTLVMAKKGVPVLNK-SNMRGDQLVRVQVEIPKRLSKEEKKLIEEL 377
> At4g39960
Length=396
Score = 123 bits (308), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 75/239 (31%), Positives = 125/239 (52%), Gaps = 7/239 (2%)
Query 25 GLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTR---RTSPCPVCEGQGMQT 81
G D + L+F +A + + R E+C C G + + + C C GQG
Sbjct 149 GEDEYYSLILNFKEAVF--GIEKEIEISRLESCGTCNGSGAKAGTKPTKCKTCGGQGQVV 206
Query 82 ETRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLR 141
+ R+ G +TCS CN TG P C +C G+G + + + + +PAGV GS LR
Sbjct 207 ASTRTPLGVFQQVMTCSPCNGTGEISKP-CGACSGDGRVRRTKRISLKVPAGVDSGSRLR 265
Query 142 VEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDG 201
V GEG+ G+ G + GDL+ I V L R+ ++ IS DA+ G ++ V TVDG
Sbjct 266 VRGEGNAGKRGGSPGDLFAVIEVIPDPVLKRDDTNILYTCKISYVDAILGTTLKVPTVDG 325
Query 202 DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSL 260
+V+LK+PA ++ ++ +G ++ S RGD +V ++V +P+ + +++ L++ L
Sbjct 326 EVDLKVPAGTQPSTTLVMAKKGVPVLNK-SKMRGDQLVRVQVEIPKRLSKEEKMLVEEL 383
> At3g17830
Length=493
Score = 117 bits (292), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 72/239 (30%), Positives = 125/239 (52%), Gaps = 10/239 (4%)
Query 26 LDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAG---PSTRRTSPCPVCEGQGMQTE 82
LD+ + LSF +A + V ETC C G S+ C C+G+G
Sbjct 183 LDIRYDLRLSFEEAVF--GVKREIEVSYLETCDGCGGTGAKSSNSIKQCSSCDGKGRVMN 240
Query 83 TRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRV 142
++R+ FG ++ TCS C G + C+ C G G + + V +P GV + + +R+
Sbjct 241 SQRTPFGIMSQVSTCSKCGGEGKTITDKCRKCIGNGRLRARKKMDVVVPPGVSDRATMRI 300
Query 143 EGEGS-EGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDG 201
+GEG+ + R+G A GDL+I + V+ + REG ++ S +I DA+ G + V TV+G
Sbjct 301 QGEGNMDKRSGRA-GDLFIVLQVDEKRGIRREGLNLYSNINIDFTDAILGATTKVETVEG 359
Query 202 DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSL 260
++L+IP ++ G + +G D PS RGDH +++++P+ ++R+L++
Sbjct 360 SMDLRIPPGTQPGDTVKLPRKGVPDTDRPS-IRGDHCFVVKISIPK--KLRERKLVEEF 415
> SPCC4G3.14
Length=528
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 73/252 (28%), Positives = 129/252 (51%), Gaps = 21/252 (8%)
Query 25 GLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGMQ 80
G D+EA +T+ F++A L TC++C G P + +++ C C+G G +
Sbjct 210 GEDIEASITIDFMEAVRGAKKDL--SYSVSSTCSSCHGSGLQPGSHKST-CFACKGTGQR 266
Query 81 TETRRSSFGFVTTSVTCSACNATGISQGP--LCKSCEGEGLISKDVHLKVNIPAGVLEGS 138
SF TT C +C TG + P C+SC G G + + + ++IP G+ + +
Sbjct 267 LHFIPPSFHMQTT---CDSCGGTGTTIPPNSACRSCMGSGTVRERKTVSIDIPPGIDDNT 323
Query 139 LLRVEGEGSE------GRNGEAR-GDLYISIGVESHDRLTREGADVCSEESISLEDAVYG 191
+LRV G G++ G N ++R GDL+ +I V H REG +V I + A G
Sbjct 324 VLRVMGAGNDASTAKGGPNAKSRPGDLFATIHVRKHPFFVREGTNVTYNAKIPMTTAALG 383
Query 192 CSITVHTVDGDVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATA 251
++ V T+ G+V+L++ + +G + + G+G +K++ + G+ V V +P+ +
Sbjct 384 GTLRVPTLTGNVDLRVSPGTSTGDRITMAGKGIRKVN--TSRYGNFYVNFEVTIPKILSP 441
Query 252 KDRELLKSLREA 263
+R LL+ L +A
Sbjct 442 HERSLLEQLADA 453
> CE27991
Length=446
Score = 95.5 bits (236), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 70/244 (28%), Positives = 117/244 (47%), Gaps = 21/244 (8%)
Query 29 EAHVTLSFVDAALNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGMQTETR 84
E + +SF + A+ GA V V E C C G P ++TS CP C G G ++
Sbjct 165 EMVMDISF-EEAVRGATK-NVSVNVVEDCLKCHGTQVEPGHKKTS-CPYCNGTGAVSQRL 221
Query 85 RSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEG 144
+ F + TT C+ C +G C+ CEGEG + + N+PAG G L+ +
Sbjct 222 QGGFFYQTT---CNRCRGSGHYNKNPCQECEGEGQTVQRRQVSFNVPAGTNNGDSLKFQ- 277
Query 145 EGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGDVE 204
G+N L++ V + RE D+ + ISL AV G ++ V ++GD
Sbjct 278 ---VGKN-----QLFVRFNVAPSLKFRREKDDIHCDVDISLAQAVLGGTVKVPGINGDTY 329
Query 205 LKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSLREAE 264
+ IPA + S K + G+G K++ S GD + ++V +P+ TA+ ++++ + E
Sbjct 330 VHIPAGTGSHTKMRLTGKGVKRLH--SYGNGDQYMHIKVTVPKYLTAEQKQIMLAWAATE 387
Query 265 DVGE 268
+ +
Sbjct 388 QLKD 391
> CE24911
Length=456
Score = 95.1 bits (235), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 70/242 (28%), Positives = 116/242 (47%), Gaps = 21/242 (8%)
Query 29 EAHVTLSFVDAALNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGMQTETR 84
E + +SF + A+ GA V V E C C G P ++TS CP C G G ++
Sbjct 165 EMVMDISF-EEAVRGATK-NVSVNVVEDCLKCHGTQVEPGHKKTS-CPYCNGTGAVSQRL 221
Query 85 RSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEG 144
+ F + TT C+ C +G C+ CEGEG + + N+PAG G L+ +
Sbjct 222 QGGFFYQTT---CNRCRGSGHYNKNPCQECEGEGQTVQRRQVSFNVPAGTNNGDSLKFQ- 277
Query 145 EGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGDVE 204
G+N L++ V + RE D+ + ISL AV G ++ V ++GD
Sbjct 278 ---VGKN-----QLFVRFNVAPSLKFRREKDDIHCDVDISLAQAVLGGTVKVPGINGDTY 329
Query 205 LKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSLREAE 264
+ IPA + S K + G+G K++ S GD + ++V +P+ TA+ ++++ + E
Sbjct 330 VHIPAGTGSHTKMRLTGKGVKRLH--SYGNGDQYMHIKVTVPKYLTAEQKQIMLAWAATE 387
Query 265 DV 266
+
Sbjct 388 QL 389
> At5g48030
Length=461
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 72/254 (28%), Positives = 113/254 (44%), Gaps = 28/254 (11%)
Query 25 GLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGMQ 80
G DV+ + LSF++A CS V + + C C G P T+R C C G GM
Sbjct 208 GQDVKVLLDLSFMEAVQ--GCSKTVTFQTEMACNTCGGQGVPPGTKREK-CKACNGSGMF 264
Query 81 TETRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLL 140
+ + G+V +CKSC G ++ +KV I GV L
Sbjct 265 HFSDLTEEGYVKHPNNLPES---------ICKSCRGARVVRGQKSVKVTIDPGVDNSDTL 315
Query 141 RVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLED----------AVY 190
+V G G+ GDLY+++ V REG+D+ + +S+ A+
Sbjct 316 KVARVGGADPEGDQPGDLYVTLKVREDPVFRREGSDIHVDAVLSVTQHLFWTSGAVSAIL 375
Query 191 GCSITVHTVDGDVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETAT 250
G +I V T+ GDV +K+ ++ G K +++ +G + S GD V V++P T
Sbjct 376 GGTIQVPTLTGDVVVKVRPGTQPGHKVVLRNKGIRA--RKSTKFGDQYVHFNVSIPANIT 433
Query 251 AKDRELLKSLREAE 264
+ RELL+ +AE
Sbjct 434 QRQRELLEEFSKAE 447
> At1g28210
Length=427
Score = 92.0 bits (227), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 66/252 (26%), Positives = 110/252 (43%), Gaps = 22/252 (8%)
Query 20 QGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAG---PSTRRTSPCPVCEG 76
+ +Q D+ ++LS +AA C+ + C +C G PS S CP C G
Sbjct 151 ENNQIKPDIRVELSLSLSEAA--EGCTKRLSFDAYVFCDSCDGLGHPSDAAMSICPTCRG 208
Query 77 QGMQTETRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLE 136
G T + +C C TG C SC G G++ ++ IP GV
Sbjct 209 VGRVT--------IPPFTASCQTCKGTGHIIKEYCMSCRGSGIVEGTKTAELVIPGGVES 260
Query 137 GSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITV 196
+ + + G G+ G+LYI + V + TR+G+D+ + +IS A+ G + V
Sbjct 261 EATITIVGAGNVSSRTSQPGNLYIKLKVANDSTFTRDGSDIYVDANISFTQAILGGKVVV 320
Query 197 HTVDGDVELKIPALSRSGRKFLIKGRGAKK----IDDPSGARGDHVVTLRVALPETATAK 252
T+ G ++L IP ++ + +++G+G K +D GD V RV P +
Sbjct 321 PTLSGKIQLDIPKGTQPDQLLVLRGKGLPKQGFFVD-----HGDQYVRFRVNFPTEVNER 375
Query 253 DRELLKSLREAE 264
R +L+ + E
Sbjct 376 QRAILEEFAKEE 387
> 7295145
Length=400
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 60/243 (24%), Positives = 116/243 (47%), Gaps = 23/243 (9%)
Query 29 EAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTRR------TSPCPVCEGQGMQTE 82
E + L F++A + C + +R C C G S PC C G G +
Sbjct 158 EFDLPLDFLEATV--GCKKRIELRYLRKCETCKGKSQLMAHRDVGKEPCRRCNGTG-KVM 214
Query 83 TRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRV 142
T+ +F V T C+ C + C++C G + +V + V++P+G +G ++ +
Sbjct 215 TKTPTFSSVNT---CTQCKGKRFTNRNDCETCSNRGFVVSNVDVMVSVPSGSRDGDVVNI 271
Query 143 EGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGD 202
N E + + + V S D R G D+ +++ +++ +A+ G S + +
Sbjct 272 I-------NPETKQQVTYRLSVPSSDYFRRVGNDILTDKHLNISEAILGGSFQIRGLYES 324
Query 203 VELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSLRE 262
VEL++ ++S + ++ G+G + + G+H+VTL+V +P + K R+L+ +L +
Sbjct 325 VELRVEPGTQSHTQVVLNGKGVRSRE----GVGNHIVTLKVRIPRNLSVKQRQLVLALSQ 380
Query 263 AED 265
AED
Sbjct 381 AED 383
> 7291628
Length=506
Score = 86.3 bits (212), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 63/205 (30%), Positives = 100/205 (48%), Gaps = 21/205 (10%)
Query 57 CTACAG----PSTRRTSPCPVCEGQGMQTETRRSSFGFVTTSVTCSACNATGISQGPLCK 112
C CAG P T+ C C G G +T S+ FV S TC C T C
Sbjct 226 CPKCAGTKCEPGTK-PGRCQYCNGTGFETV---STGPFVMRS-TCRYCQGTRQHIKYPCS 280
Query 113 SCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTR 172
CEG+G + + V +PAG+ G +R++ E L+++ VE D R
Sbjct 281 ECEGKGRTVQRRKVTVPVPAGIENGQTVRMQVGSKE---------LFVTFRVERSDYFRR 331
Query 173 EGADVCSEESISLEDAVYGCSITVHTVDGDVELKIPALSRSGRKFLIKGRGAKKIDDPSG 232
EGADV ++ +ISL AV G ++ V V D + + + S K +++G+G K+++ +
Sbjct 332 EGADVHTDAAISLAQAVLGGTVRVQGVYEDQWINVEPGTSSHHKIMLRGKGLKRVN--AH 389
Query 233 ARGDHVVTLRVALPETATAKDRELL 257
GDH V +++ +P +A D++ L
Sbjct 390 GHGDHYVHVKITVP-SAKKLDKKRL 413
> ECU07g0760
Length=398
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 56/178 (31%), Positives = 80/178 (44%), Gaps = 5/178 (2%)
Query 51 VRRQETCTACAGPSTRRTSPCPVCEGQGMQTETRRSSFGFVTTSVT-CSACNATGIS-QG 108
VR ++ CT C G + C C G G+ T +RRS GFVT + T C C+ +G +G
Sbjct 146 VRTEKVCTTCDGKGGKDVETCKKCNGNGVYT-SRRSLGGFVTLAETRCDGCDGSGHKIKG 204
Query 109 PLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHD 168
C +C G I +VNI GV +G + EG G + R G GD+ I V+
Sbjct 205 KPCSTCNGAEYIQDKTMFEVNIKPGVRKGEKIVFEGMGDQ-RRGHVPGDVIFIIDVQEDS 263
Query 169 RLTREGADVCSEESISLEDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRGAK 225
R R G D+ I L A+ G + +DG +E+ + I+ G K
Sbjct 264 RFERCGNDLVGNIDIPLYTAIGGGVVYFTHIDGRQLEINVSPFRTFDTALKIRNEGFK 321
> Hs4827026
Length=480
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 62/242 (25%), Positives = 114/242 (47%), Gaps = 26/242 (10%)
Query 29 EAHVTLSFVDAA--LNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGMQTE 82
E + L+F AA +N ++ ++ +TC C G P T+ C C G GM+T
Sbjct 210 EYFMELTFNQAAKGVNKEFTVNIM----DTCERCNGKGNEPGTK-VQHCHYCGGSGMETI 264
Query 83 TRRSSFGFVTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRV 142
++ FV S TC C G C C G G + + + +PAGV +G +R+
Sbjct 265 ---NTGPFVMRS-TCRRCGGRGSIIISPCVVCRGAGQAKQKKRVMIPVPAGVEDGQTVRM 320
Query 143 EGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGD 202
+ +++I+ V+ R+GAD+ S+ IS+ A+ G + +
Sbjct 321 P---------VGKREIFITFRVQKSPVFRRDGADIHSDLFISIAQALLGGTARAQGLYET 371
Query 203 VELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSLRE 262
+ + IP +++ +K + G+G +I+ S GDH + +++ +P+ T++ + L+ S E
Sbjct 372 INVTIPPGTQTDQKIRMGGKGIPRIN--SYGYGDHYIHIKIRVPKRLTSRQQSLILSYAE 429
Query 263 AE 264
E
Sbjct 430 DE 431
> YFL016c
Length=511
Score = 72.8 bits (177), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 64/256 (25%), Positives = 120/256 (46%), Gaps = 28/256 (10%)
Query 24 QGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAG----PSTRRTSPCPVCEGQGM 79
+G +E +SF DA G+ ++ + + C+ C+G P+T + S C C G G
Sbjct 198 RGDPIEIVHKVSFKDAVF-GSKNVQLRFSALDPCSTCSGTGMKPNTHKVS-CSTCHGTGT 255
Query 80 QTETRRSSFGFVTTSVTCSACNATGISQGPL--CKSCEGEGL-ISKDVHLKVNIPAGVLE 136
R GF S TC CN G + P C C GEG+ +++ + V++P G+ +
Sbjct 256 TVHIRG---GFQMMS-TCPTCNGEGTMKRPQDNCTKCHGEGVQVNRAKTITVDLPHGLQD 311
Query 137 GSLLRVEGEGS----------EGRNGEARGDLYISIGVESHDRLT-REGADVCSEESISL 185
G ++R+ G+GS + +RGD+ + I V+ + + D+ ++ I +
Sbjct 312 GDVVRIPGQGSYPDIAVEADLKDSVKLSRGDILVRIRVDKDPNFSIKNKYDIWYDKEIPI 371
Query 186 EDAVYGCSITVHTVDGD-VELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVA 244
A G ++T+ TV+G + +K+ ++ + I G K S RGD V ++
Sbjct 372 TTAALGGTVTIPTVEGQKIRIKVAPGTQYNQVISIPNMGVPKT---STIRGDMKVQYKIV 428
Query 245 LPETATAKDRELLKSL 260
+ + + ++ L ++L
Sbjct 429 VKKPQSLAEKCLWEAL 444
> YNL064c
Length=409
Score = 72.8 bits (177), Expect = 7e-13, Method: Compositional matrix adjust.
Identities = 65/259 (25%), Positives = 114/259 (44%), Gaps = 17/259 (6%)
Query 13 AAGRRHTQGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTRR--TSP 70
A G + +G Q+G D++ ++ S + L + + + +Q C C G ++
Sbjct 101 AGGAQRPRGPQRGKDIKHEISASLEE--LYKGRTAKLALNKQILCKECEGRGGKKGAVKK 158
Query 71 CPVCEGQGMQTETRRSSFGFVTTSVTCSACNATG--ISQGPLCKSCEGEGLISKDVHLKV 128
C C GQG++ TR+ C C+ TG I CKSC G+ + ++ L+V
Sbjct 159 CTSCNGQGIKFVTRQMGPMIQRFQTECDVCHGTGDIIDPKDRCKSCNGKKVENERKILEV 218
Query 129 NIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDA 188
++ G+ +G + +GE + + GD+ + H R+G D+ E I L A
Sbjct 219 HVEPGMKDGQRIVFKGEADQAPD-VIPGDVVFIVSERPHKSFKRDGDDLVYEAEIDLLTA 277
Query 189 VYGCSITVHTVDGDVELKIPALS----RSGRKFLIKGRGAKKIDDPS-GARGDHVVTLRV 243
+ G + V GD LK+ + G + +I+G+G + P G G+ ++ +
Sbjct 278 IAGGEFALEHVSGDW-LKVGIVPGEVIAPGMRKVIEGKG---MPIPKYGGYGNLIIKFTI 333
Query 244 ALPETATAKDRELLKSLRE 262
PE + E LK L E
Sbjct 334 KFPENHFTSE-ENLKKLEE 351
> Hs5031741
Length=412
Score = 70.5 bits (171), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 54/200 (27%), Positives = 93/200 (46%), Gaps = 10/200 (5%)
Query 57 CTACAGPSTRR--TSPCPVCEGQGMQTETRRSSFGFVT-TSVTCSACNATG--ISQGPLC 111
C+AC+G + C C G+G++ R+ + G V CS CN G I++ C
Sbjct 143 CSACSGQGGKSGAVQKCSACRGRGVRIMIRQLAPGMVQQMQSVCSDCNGEGEVINEKDRC 202
Query 112 KSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLT 171
K CEG+ +I + L+V++ G+ G + GE + E GD+ + + + H+
Sbjct 203 KKCEGKKVIKEVKILEVHVDKGMKHGQRITFTGEADQAPGVEP-GDIVLLLQEKEHEVFQ 261
Query 172 REGADVCSEESISLEDAVYGCSITVHTVDG-DVELKIP--ALSRSGRKFLIKGRGAKKID 228
R+G D+ I L +A+ G T +DG + +K P + G +++G G +
Sbjct 262 RDGNDLHMTYKIGLVEALCGFQFTFKHLDGRQIVVKYPPGKVIEPGCVRVVRGEGMPQYR 321
Query 229 DPSGARGDHVVTLRVALPET 248
+P +GD + V PE
Sbjct 322 NPF-EKGDLYIKFDVQFPEN 340
> Hs6631085
Length=337
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 42/147 (28%), Positives = 75/147 (51%), Gaps = 3/147 (2%)
Query 113 SCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTR 172
+ +G S+D L + I G EG+ + EG E N D+ I + H + R
Sbjct 187 NADGRSYRSEDKILTIEIKKGWKEGTKITFPREGDETPNS-IPADIVFIIKDKDHPKFKR 245
Query 173 EGADVCSEESISLEDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRGAKKIDDPS 231
+G+++ ISL +A+ GCSI V T+DG ++ + + + + G + I G G +P
Sbjct 246 DGSNIIYTAKISLREALCGCSINVPTLDGRNIPMSVNDIVKPGMRRRIIGYGLPFPKNPD 305
Query 232 GARGDHVVTLRVALPETATAKDRELLK 258
RGD ++ V+ P+T ++ +E+L+
Sbjct 306 -QRGDLLIEFEVSFPDTISSSSKEVLR 331
> SPBC1347.05c
Length=386
Score = 65.5 bits (158), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 53/197 (26%), Positives = 88/197 (44%), Gaps = 13/197 (6%)
Query 15 GRRHTQGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGP-------STRR 67
GRR ++G +E V + G+ +L + V+R TC+ C+G + +
Sbjct 111 GRRRQNAVRRGPSMEQIVQIHLSSFYTGGSFTLEIPVKR--TCSVCSGQGFNPKYSADKA 168
Query 68 TSPCPVCEGQGMQTETRRSSFGF-VTTSVTCSACNATGISQGPLCKSCEGEGLISKDVHL 126
CPVC G G + + GF + C+ACN G + C C+GE +
Sbjct 169 IESCPVCGGSGFRVIEHMIAPGFRQQMRMPCNACNGNGRTIKHKCPRCKGERVAEVVESF 228
Query 127 KVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDR-LTREGADVCSEESISL 185
+ +PAG EG + G+ E EA GD+ + + D TR+ D+ +E+IS+
Sbjct 229 DIKVPAGAPEGYRIGFRGKADEIPGMEA-GDIIVILEAAGGDYGWTRKDNDLYRKETISV 287
Query 186 EDAVYG-CSITVHTVDG 201
+A+ G + +DG
Sbjct 288 REALLGNWKRKIQKLDG 304
> CE16015
Length=402
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/155 (27%), Positives = 69/155 (44%), Gaps = 4/155 (2%)
Query 51 VRRQETCTACAGPSTRRTS--PCPVCEGQGMQTETRRSSFGFVTTSVTCSACNATG--IS 106
+ R TC C G S C C G+G++ R C +CN G
Sbjct 130 ISRTATCKGCKGLGGNEGSAKECSDCRGRGIKVRVIRMGPMVQQMQSHCDSCNGEGSTFL 189
Query 107 QGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVES 166
+ CK C G+ + +D ++V I G+ +G EG+G E E GD + +
Sbjct 190 EKDRCKKCNGKKQVKEDEIIEVGITPGMKDGEKFVFEGKGDEVIGIEKPGDFVVVLDEVE 249
Query 167 HDRLTREGADVCSEESISLEDAVYGCSITVHTVDG 201
H++ R+G ++ + +I L +A+ G T+ T+DG
Sbjct 250 HEKFVRKGDNLIIQHNIDLSEALCGFVRTISTLDG 284
> Hs7706495
Length=358
Score = 64.7 bits (156), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 48/158 (30%), Positives = 77/158 (48%), Gaps = 4/158 (2%)
Query 105 ISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGV 164
++Q +C C L++++ L+V I GV +G GEG +GE GDL I V
Sbjct 187 MTQEVVCDECPNVKLVNEERTLEVEIEPGVRDGMEYPFIGEGEPHVDGEP-GDLRFRIKV 245
Query 165 ESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRG 223
H R G D+ + +ISL +++ G + + +DG V + ++R G K KG G
Sbjct 246 VKHPIFERRGDDLYTNVTISLVESLVGFEMDITHLDGHKVHISRDKITRPGAKLWKKGEG 305
Query 224 AKKIDDPSGARGDHVVTLRVALP-ETATAKDRELLKSL 260
D+ + +G ++T V P E T + RE +K L
Sbjct 306 LPNFDN-NNIKGSLIITFDVDFPKEQLTEEAREGIKQL 342
> At3g44110
Length=420
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 47/185 (25%), Positives = 84/185 (45%), Gaps = 8/185 (4%)
Query 22 SQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTRR--TSPCPVCEGQGM 79
++G DV + +S D L L + R C+ C G ++ + C C+G GM
Sbjct 115 QRRGEDVVHPLKVSLEDVYLGTMKKLSL--SRNALCSKCNGKGSKSGASLKCGGCQGSGM 172
Query 80 QTETRRSSFGFVT-TSVTCSACNATG--ISQGPLCKSCEGEGLISKDVHLKVNIPAGVLE 136
+ R+ G + C+ C TG I+ C C+G+ +I + L+VN+ G+
Sbjct 173 KVSIRQLGPGMIQQMQHACNECKGTGETINDRDRCPQCKGDKVIPEKKVLEVNVEKGMQH 232
Query 137 GSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITV 196
+ EG+ E + GD+ + + H + R+G D+ E ++SL +A+ G +
Sbjct 233 SQKITFEGQADEAPDT-VTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFVL 291
Query 197 HTVDG 201
+DG
Sbjct 292 THLDG 296
> CE13229
Length=439
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 53/218 (24%), Positives = 92/218 (42%), Gaps = 19/218 (8%)
Query 57 CTACAGPSTRR--TSPCPVCEGQGMQTETRRSSFGFVT-TSVTCSACNATG--ISQGPLC 111
C C G ++ C C G+G++T ++ G + V C AC +G + G C
Sbjct 169 CKTCEGSGGKKGEKYKCDACRGRGVKTIVQQIGPGMLQQMQVHCDACKGSGGKVPAGDKC 228
Query 112 KSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLT 171
K C GE + L+V++ G+ + +G+G + GD+ I I + HD
Sbjct 229 KGCHGEKYENVSKILEVHVLPGMKHNDKITFKGDGDQSDPDGEPGDVVIVIQQKDHDIFK 288
Query 172 REGADVCSEESISLEDAVYGCSITVHTVDGDVELKIPALSRSGRKFLIK--------GRG 223
R+G D+ + +SL +A+ G + + +DG P + S + +IK G+G
Sbjct 289 RDGDDLHMTKKLSLNEALCGYNFLIKHLDGH-----PLVLSSKQGDVIKPGVIRGVLGKG 343
Query 224 AKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSLR 261
P +G+ V V P+ D + L+
Sbjct 344 MPNKKYPE-LKGNLFVEFEVEFPKEHFLDDEKAYAVLK 380
> Hs4504511
Length=397
Score = 63.5 bits (153), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 52/188 (27%), Positives = 84/188 (44%), Gaps = 8/188 (4%)
Query 19 TQGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTRR--TSPCPVCEG 76
Q ++G +V ++++ D NGA + + +++ C C G ++ CP C G
Sbjct 98 MQRERRGKNVVHQLSVTLEDL-YNGA-TRKLALQKNVICDKCEGRGGKKGAVECCPNCRG 155
Query 77 QGMQTETRRSSFGFVT-TSVTCSACNATG--ISQGPLCKSCEGEGLISKDVHLKVNIPAG 133
GMQ + G V C C G IS CKSC G ++ + L+V+I G
Sbjct 156 TGMQIRIHQIGPGMVQQIQSVCMECQGHGERISPKDRCKSCNGRKIVREKKILEVHIDKG 215
Query 134 VLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCS 193
+ +G + GEG + G GD+ I + + H TR G D+ I L +A+ G
Sbjct 216 MKDGQKITFHGEGDQ-EPGLEPGDIIIVLDQKDHAVFTRRGEDLFMCMDIQLVEALCGFQ 274
Query 194 ITVHTVDG 201
+ T+D
Sbjct 275 KPISTLDN 282
> YMR214w
Length=404
Score = 63.2 bits (152), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 61/239 (25%), Positives = 108/239 (45%), Gaps = 23/239 (9%)
Query 23 QQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTAC--AGPSTRRTSPCPVCEGQGMQ 80
Q+G ++ LS S+ + + C AC +G + + + CP C+G+G+
Sbjct 164 QRGPMIKVQEKLSL--KQFYSGSSIEFTLNLNDECDACHGSGSADGKLAQCPDCQGRGVI 221
Query 81 TETRRSSFGFVTTSV--TCSACNATGISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGS 138
+ R G +T + C C TG CK+C G+ + K+ V++P G
Sbjct 222 IQVLR--MGIMTQQIQQMCGRCGGTGQIIKNECKTCHGKKVTKKNKFFHVDVPPGAPRNY 279
Query 139 LLRVEGEGSEGRNGEARGDLYISIGVESHDRL--TREGADVCSEESISLEDAVY-GCSIT 195
+ GE +G + +A GDL I + + + R G ++ E +S +A+Y G T
Sbjct 280 MDTRVGEAEKGPDFDA-GDLVIEFKEKDTENMGYRRRGDNLYRTEVLSAAEALYGGWQRT 338
Query 196 VHTVDGD--VELKIPA--LSRSGRKFLIKGRGAKKIDDPSGAR--GDHVVTLRVALPET 248
+ +D + V+L PA + +G ++KG G P G++ GD + V +P+T
Sbjct 339 IEFLDENKPVKLSRPAHVVVSNGEVEVVKGFGM-----PKGSKGYGDLYIDYVVVMPKT 392
> 7296201
Length=354
Score = 62.8 bits (151), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 42/156 (26%), Positives = 78/156 (50%), Gaps = 4/156 (2%)
Query 107 QGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVES 166
Q +C C L++++ L++ + G+++G R EG +GE GDL + +
Sbjct 188 QQTVCDECPNVKLVNEERTLEIEVEQGMVDGQETRFVAEGEPHIDGEP-GDLIVRVQQMP 246
Query 167 HDRLTREGADVCSEESISLEDAVYGCSITVHTVDGD-VELKIPALSRSGRKFLIKGRGAK 225
H R R+ D+ + +ISL+DA+ G S+ + +DG V + ++ G + KG G
Sbjct 247 HPRFLRKNDDLYTNVTISLQDALVGFSMEIKHLDGHLVPVTREKVTWPGARIRKKGEGMP 306
Query 226 KIDDPSGARGDHVVTLRVALPET-ATAKDRELLKSL 260
++ + G+ +T V P+ T +D+E LK +
Sbjct 307 NFEN-NNLTGNLYITFDVEFPKKDLTEEDKEALKKI 341
> SPBC1734.11
Length=407
Score = 62.4 bits (150), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 61/245 (24%), Positives = 102/245 (41%), Gaps = 14/245 (5%)
Query 20 QGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTRRTS--PCPVCEGQ 77
+G ++G D+ + ++ D L + + ++++ C C+G + S C C G
Sbjct 102 RGPRKGKDLVHTIKVTLED--LYRGKTTKLALQKKVICPKCSGRGGKEGSVKSCASCNGS 159
Query 78 GMQTETRRSSFGFVTTSVTCSACNATG--ISQGPLCKSCEGEGLISKDVHLKVNIPAGVL 135
G++ TR +TC CN G I CK C+G +IS+ L V++ G+
Sbjct 160 GVKFITRAMGPMIQRMQMTCPDCNGAGETIRDEDRCKECDGAKVISQRKILTVHVEKGMH 219
Query 136 EGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSIT 195
G + + EG + G GD+ I + H R R G + E + L A+ G I
Sbjct 220 NGQKIVFKEEGEQA-PGIIPGDVIFVIDQKEHPRFKRSGDHLFYEAHVDLLTALAGGQIV 278
Query 196 VHTVDGDVELKIPALS----RSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVALPETATA 251
V +D D L IP + R ++ G+G + G+ + V PE A
Sbjct 279 VEHLD-DRWLTIPIIPGECIRPNELKVLPGQGM--LSQRHHQPGNLYIRFHVDFPEPNFA 335
Query 252 KDREL 256
+L
Sbjct 336 TPEQL 340
> CE01664
Length=355
Score = 61.6 bits (148), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 41/152 (26%), Positives = 75/152 (49%), Gaps = 3/152 (1%)
Query 105 ISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGV 164
+ Q +C C L+ ++ L+V + G G GEG G+ GDL I +
Sbjct 186 MFQVKVCDECPNVKLVQENKVLEVEVEVGADNGHQQIFHGEGEPHIEGDP-GDLKFKIRI 244
Query 165 ESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGD-VELKIPALSRSGRKFLIKGRG 223
+ H R R+G D+ + +ISL+DA+ G + + +DG V+++ ++ G + K G
Sbjct 245 QKHPRFERKGDDLYTNVTISLQDALNGFEMEIQHLDGHIVKVQRDKVTWPGARLRKKDEG 304
Query 224 AKKIDDPSGARGDHVVTLRVALPETATAKDRE 255
++D + +G VVT V P+T + +++
Sbjct 305 MPSLED-NNKKGMLVVTFDVEFPKTELSDEQK 335
> YLR090w
Length=459
Score = 60.5 bits (145), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 62/265 (23%), Positives = 105/265 (39%), Gaps = 33/265 (12%)
Query 27 DVEAHVTLSFVDAALNGACSLPVIVRRQETCTACAGPSTR--------RTSPCPVCEGQG 78
D++ ++L+ D + L ++RQ C C G + C C G+G
Sbjct 131 DIDIDISLTLKDLYMGK--KLKFDLKRQVICIKCHGSGWKPKRKIHVTHDVECESCAGKG 188
Query 79 MQTETRRSSFGFVTTS-VTCSACNATG-ISQGP-----LCKSCEGEGLISKDVHLKVNIP 131
+ +R G V + V C CN G ++ P C C G GL+SK + VN+
Sbjct 189 SKERLKRFGPGLVASQWVVCEKCNGKGKYTKRPKNPKNFCPDCAGLGLLSKKEIITVNVA 248
Query 132 AGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTRE-----------GADVCSE 180
G ++ V+G E + GDL + E + L ++ G D+ +
Sbjct 249 PGHHFNDVITVKGMADEEIDKTTCGDLKFHL-TEKQENLEQKQIFLKNFDDGAGEDLYTS 307
Query 181 ESISLEDAVYGC-SITVHTVDG---DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGD 236
+ISL +A+ G T D + +K + R G I G +D+P G GD
Sbjct 308 ITISLSEALTGFEKFLTKTFDDRLLTLSVKPGRVVRPGDTIKIANEGWPILDNPHGRCGD 367
Query 237 HVVTLRVALPETATAKDRELLKSLR 261
V + + P ++ L +++
Sbjct 368 LYVFVHIEFPPDNWFNEKSELLAIK 392
> YNL077w
Length=528
Score = 60.1 bits (144), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 37/121 (30%), Positives = 56/121 (46%), Gaps = 9/121 (7%)
Query 51 VRRQETCTACAGPSTRRTSPCPVCEGQGMQTETRRSSFGFVTTSVTCSACNATG--ISQG 108
+ R C+ C G + C C+GQG+QT+TRR + S TC+ C G +
Sbjct 200 LNRTRICSVCDGHGGLKKCTCKTCKGQGIQTQTRRMGPLVQSWSQTCADCGGAGVFVKNK 259
Query 109 PLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSE-------GRNGEARGDLYIS 161
+C+ C+G G I + L+V + G L+ + GEG E G GD+ I+
Sbjct 260 DICQQCQGLGFIKERKILQVTVQPGSCHNQLIVLTGEGDEVISTKGGGHEKVIPGDVVIT 319
Query 162 I 162
I
Sbjct 320 I 320
> SPBC405.06
Length=413
Score = 59.7 bits (143), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 54/205 (26%), Positives = 88/205 (42%), Gaps = 12/205 (5%)
Query 53 RQETCTACAGPSTRRTS---PCPVCEGQGMQTETRRSSFGFVTTS-VTCSACNATGISQ- 107
R C C G +R + PC C+G+G++ + VT S V C CN G+S
Sbjct 143 RNTLCPRCQGRGGKRFAKEKPCLSCDGKGVKQHLKHVGPHHVTNSQVICDTCNGKGVSFR 202
Query 108 -GPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVES 166
CK C+G G + + L + E + G E G GD+ + + +
Sbjct 203 GKDRCKHCKGSGTVPEQRMLSFFVNRSAKENDKIIQRGMADEAY-GITPGDVILQLHQKP 261
Query 167 HDRLTREGADVCSEESISLEDAVYGCS-ITVHTVDG---DVELKIPALSRSGRKFLIKGR 222
H R G D+ ++ ISL +A+ G + + + T+DG + I + G +I G
Sbjct 262 HPVFERLGDDLKAKLKISLAEALTGFNRVILTTLDGRGLEYVQPIGKILHPGDCLIIPGE 321
Query 223 GAKKIDDPSGARGDHVVTLRVALPE 247
G K D + RGD + + + P+
Sbjct 322 GMYK-DSKTDLRGDLYLEVDIEFPK 345
> At1g44160
Length=352
Score = 57.0 bits (136), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 44/164 (26%), Positives = 76/164 (46%), Gaps = 15/164 (9%)
Query 110 LCKSCEGEGLISKDV------------HLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGD 157
LC C + I +DV +++ + G G+ + EG+G+E D
Sbjct 186 LCNGCTKKIKIKRDVITSLGEKCEEEEMVEIKVKPGWKGGTKVTFEGKGNEAMRS-VPAD 244
Query 158 LYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGD-VELKIPALSRSGRK 216
L I + H+ REG D+ +SL +A+ GC ++V +DGD + L+I + G
Sbjct 245 LTFVIVEKEHEVFKREGDDLEMAVEVSLLEALTGCELSVALLDGDNMRLRIEDVIHPGYV 304
Query 217 FLIKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKSL 260
+++G+G + + G RGD V R P+ T + R + S+
Sbjct 305 TVVQGKGMPNLKE-KGKRGDLRVRFRTKFPQHLTDEQRAEIHSI 347
> At1g10350
Length=349
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 41/135 (30%), Positives = 67/135 (49%), Gaps = 6/135 (4%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
LK++I G +G+ + +G++ G DL + + H R+G D+ E+ +SL
Sbjct 213 LKIDIKPGWKKGTKITFPEKGNQ-EPGVTPADLIFVVDEKPHSVFKRDGNDLILEKKVSL 271
Query 186 EDAVYGCSITVHTVDGDVELKIPALS--RSGRKFLIKGRGAKKIDDPSGARGDHVVTLRV 243
DA+ G +I+V T+DG L IP L + G++ +I G D RGD VT +
Sbjct 272 IDALTGLTISVTTLDGR-SLTIPVLDIVKPGQEIVIPNEGMPTKD--PLKRGDLRVTFEI 328
Query 244 ALPETATAKDRELLK 258
P T++ + LK
Sbjct 329 LFPSRLTSEQKNDLK 343
> At5g25530
Length=347
Score = 56.2 bits (134), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 33/134 (24%), Positives = 68/134 (50%), Gaps = 3/134 (2%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L + + G +G+ ++ +G+E N + DL I + HD TR+G D+ + ++L
Sbjct 211 LTIVVKPGWKKGTKIKFPDKGNEQVN-QLPADLVFVIDEKPHDLFTRDGNDLITSRRVTL 269
Query 186 EDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVA 244
+A+ G ++ ++T+DG ++ + + + G +F++ G G +P +GD + V
Sbjct 270 AEAIGGTTVNINTLDGRNLPVGVAEIVSPGYEFVVPGEGMPIAKEPRN-KGDLKIKFDVQ 328
Query 245 LPETATAKDRELLK 258
P T + + LK
Sbjct 329 FPARLTTEQKSALK 342
> CE03412
Length=331
Score = 55.8 bits (133), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 39/133 (29%), Positives = 61/133 (45%), Gaps = 3/133 (2%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L V I G G+ + EG + N D+ I + H + REG+D+ E ISL
Sbjct 197 LTVTIKPGWKSGTKITFPKEGDQHPN-RTPADIVFVIKDKPHPKFKREGSDIKRVEKISL 255
Query 186 EDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVA 244
+ A+ G I + T+DG D L++ + + G + G+G PS RGD ++ V
Sbjct 256 KSALTGLDIMIPTLDGADYRLQLNDVIKPGTTRRLTGKGLPNPKSPS-HRGDLIIEFDVE 314
Query 245 LPETATAKDRELL 257
P RE++
Sbjct 315 FPSQLNPTQREVI 327
> Hs5453690
Length=340
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 38/149 (25%), Positives = 70/149 (46%), Gaps = 3/149 (2%)
Query 113 SCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTR 172
+ +G+ + ++D L + + G EG+ + EG + N D+ + + H+ R
Sbjct 191 NPDGKSIRNEDKILTIEVKKGWKEGTKITFPKEGDQTSNN-IPADIVFVLKDKPHNIFKR 249
Query 173 EGADVCSEESISLEDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRGAKKIDDPS 231
+G+DV ISL +A+ GC++ V T+DG + + + R G + + G G P
Sbjct 250 DGSDVIYPARISLREALCGCTVNVPTLDGRTIPVVFKDVIRPGMRRKVPGEGLPLPKTPE 309
Query 232 GARGDHVVTLRVALPETATAKDRELLKSL 260
RGD ++ V PE R +L+ +
Sbjct 310 -KRGDLIIEFEVIFPERIPQTSRTVLEQV 337
> HsM6912422
Length=348
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 73/149 (48%), Gaps = 5/149 (3%)
Query 113 SCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTR 172
+ +G + ++D L + I G EG+ + EG + D+ + + H R
Sbjct 198 NPDGRTVRTEDKILHIVIKRGWKEGTKITFPKEG-DATPDNIPADIVFVLKDKPHAHFRR 256
Query 173 EGADVCSEESISLEDAVYGCSITVHTVDGDVELKIP--ALSRSGRKFLIKGRGAKKIDDP 230
+G +V ISL++A+ GC++ + T+DG V + +P + + G ++G G P
Sbjct 257 DGTNVLYSALISLKEALCGCTVNIPTIDGRV-IPLPCNDVIKPGTVKRLRGEGLPFPKVP 315
Query 231 SGARGDHVVTLRVALPETATAKDRELLKS 259
+ RGD +V +V P+ T + R++LK
Sbjct 316 T-QRGDLIVEFKVRFPDRLTPQTRQILKQ 343
> 7299759
Length=403
Score = 52.8 bits (125), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 42/169 (24%), Positives = 77/169 (45%), Gaps = 7/169 (4%)
Query 37 VDAALNGACSLPVIVRRQETCTACAGPSTRRTS--PCPVCEGQGMQTETRRSSFGFVT-T 93
++ NGA + + +++ C C G ++ S C C G G++T ++ + G +
Sbjct 121 LEELYNGA-TRKLQLQKNVICDKCEGRGGKKGSIEKCLQCRGNGVETRVQQIAPGIMQHI 179
Query 94 SVTCSACNATG--ISQGPLCKSCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRN 151
C C+ TG I + CK+C G + + L+V+I G+ +G + GEG
Sbjct 180 EQVCRKCSGTGETIQEKDRCKNCSGRKTVRERKVLEVHIEKGMRDGQKIVFTGEGDHEPE 239
Query 152 GEARGDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVD 200
+ GD+ I + + H G D+ + + L +A+ G V T+D
Sbjct 240 SQP-GDIIILLDEKEHSTFAHAGQDLMMKMPLQLVEALCGFQRIVKTLD 287
> Hs21361413
Length=348
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 39/149 (26%), Positives = 73/149 (48%), Gaps = 5/149 (3%)
Query 113 SCEGEGLISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTR 172
+ +G + ++D L + I G EG+ + EG + D+ + + H R
Sbjct 198 NPDGRTVRTEDKILHIVIKRGWKEGTKITFPKEG-DAHLDNIPADIVFVLKDKPHAHFRR 256
Query 173 EGADVCSEESISLEDAVYGCSITVHTVDGDVELKIP--ALSRSGRKFLIKGRGAKKIDDP 230
+G +V ISL++A+ GC++ + T+DG V + +P + + G ++G G P
Sbjct 257 DGTNVLYSALISLKEALCGCTVNIPTIDGRV-IPLPCNDVIKPGTVKRLRGEGLPFPKVP 315
Query 231 SGARGDHVVTLRVALPETATAKDRELLKS 259
+ RGD +V +V P+ T + R++LK
Sbjct 316 T-QRGDLIVEFKVRFPDRLTPQTRQILKQ 343
> 7298006
Length=389
Score = 52.0 bits (123), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 67/268 (25%), Positives = 112/268 (41%), Gaps = 28/268 (10%)
Query 7 PFAAAAAAGRRHTQGSQQGLDVEAHVTLSFVDAALNGACSLPVIVRRQETCTAC---AGP 63
PF ++ GR G + V+ +TL + G V RQ+ C+ C GP
Sbjct 90 PFDRVSSEGRGRRNGK---VVVKVELTL---EEIYVGGMKKKVEYNRQKLCSKCNGDGGP 143
Query 64 STRRTSPCPVCEGQGMQTETRRSSFGFVTTS---VTCSACNATG--ISQGPLCKSCEGEG 118
S C C G G R ++F F+ S TC C+ G I C C+G G
Sbjct 144 KEAHES-CETCGGAG-----RAAAFTFMGLSPFDTTCPTCDGRGFTIRDDKKCSPCQGSG 197
Query 119 LISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVC 178
+ + + + + G + EG + R GE GDL + I H R A++
Sbjct 198 FVEQKMKRDLVVERGAPHMLKVPFANEGHQMRGGEF-GDLIVVISQMEHPIFQRRHANLY 256
Query 179 SEE-SISLEDAVYGCSITVHTVDG-DVELK-IPA-LSRSGRKFLIKGRGAKKIDDPSGAR 234
+ I++ +A+ G S +DG +V L+ P + + + +++G G + + +
Sbjct 257 MRDLEINITEALCGYSHCFKHLDGRNVCLRTYPGEVLQHNQIKMVRGSGMPVFNKATDS- 315
Query 235 GDHVVTLRVALPET--ATAKDRELLKSL 260
GD + +V P+ ATA +L+ L
Sbjct 316 GDLYMKFKVKFPDNDFATAPQLAMLEDL 343
> At1g11040
Length=438
Score = 50.4 bits (119), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 41/142 (28%), Positives = 69/142 (48%), Gaps = 4/142 (2%)
Query 117 EGLI-SKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGA 175
EGLI ++ L+VNI G +G+ + EG G+E + G D+ + + H R G
Sbjct 288 EGLIMQQEEMLRVNIQPGWKKGTKITFEGVGNE-KPGYLPEDITFVVEEKRHPLFKRRGD 346
Query 176 DVCSEESISLEDAVYGCSITVHTVDGD-VELKIPALSRSGRKFLIKGRGAKKIDDPSGAR 234
D+ I L A+ GC ++V + G+ + + + + G + IKG+G + G R
Sbjct 347 DLEIAVEIPLLKALTGCKLSVPLLSGESMSITVGDVIFHGFEKAIKGQGMPNAKE-EGKR 405
Query 235 GDHVVTLRVALPETATAKDREL 256
GD +T V PE + + R +
Sbjct 406 GDLRITFLVNFPEKLSEEQRSM 427
> SPCC830.07c
Length=379
Score = 50.1 bits (118), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 35/133 (26%), Positives = 66/133 (49%), Gaps = 5/133 (3%)
Query 120 ISKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCS 179
+ D L++ + G G+ ++ GEG E +G + D+ + + H TR G D+
Sbjct 240 VKADRILEIKVKPGWKAGTKIKFAGEGDEKPDGTVQ-DIQFVLAEKPHPVFTRSGDDLRM 298
Query 180 EESISLEDAVYGCSITVHTVDGDVELKIPA--LSRSGRKFLIKGRGAKKIDDPSGARGDH 237
+ +SL++A+ G S + T+DG +LK+ + ++ G + G G +PS RG+
Sbjct 299 QVELSLKEALLGFSKQISTIDGK-KLKVSSSLPTQPGYEITYPGFGMPLPKNPS-QRGNM 356
Query 238 VVTLRVALPETAT 250
++ +V P T
Sbjct 357 IIECKVKFPTELT 369
> At1g59725
Length=331
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 36/135 (26%), Positives = 66/135 (48%), Gaps = 5/135 (3%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
LK++I G +G+ + +G++ G DL I + H R+G D+ ++ +SL
Sbjct 194 LKIDITPGWKKGTKITFPEKGNQ-EPGVTPADLIFVIDEKPHSVYKRDGNDLIVDKKVSL 252
Query 186 EDAVYGCSITVHTVDGDVELKIPALS--RSGRKFLIKGRGAKKIDDPSGARGDHVVTLRV 243
+A+ G ++++ T+DG L IP L + G++ +I G I RGD + +
Sbjct 253 LEALTGITLSLTTLDGR-NLTIPVLDIVKPGQEIVIPSEGM-PISKEGSKRGDLRINFEI 310
Query 244 ALPETATAKDRELLK 258
P T++ + LK
Sbjct 311 CFPSRLTSEQKTDLK 325
> Hs22056086
Length=354
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 54/213 (25%), Positives = 80/213 (37%), Gaps = 51/213 (23%)
Query 71 CPVCEGQGMQTETRRSSFGFVT-TSVTCSACNATG--ISQGPLCKSCEGEGLISKDVHLK 127
CP C G GMQ + G V C C G IS C+SC G ++ + L+
Sbjct 54 CPNCRGTGMQIRIHQIGPGKVQQIQSVCVECQGHGEQISPKDRCESCNGRKILREKKILE 113
Query 128 VNIPAGVLEGSLLRVEGEGSE--GRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
+I G+ G + GEG + G GE D+ I + + TR+G D
Sbjct 114 FHIDKGMKYGQKITFHGEGYQEPGLEGE---DIIIVLDQKDDAVFTRQGED--------- 161
Query 186 EDAVYGCSITVHTVDGDVELKI----PALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTL 241
++ C I H GD++ I P R K G ++
Sbjct 162 ---LFMCQIVKH---GDIKCVINEGMPIYPRPYEK------------------GRLIIEF 197
Query 242 RVALPET-ATAKDR-----ELLKSLREAEDVGE 268
+V LPE + D+ +LL +E E+ E
Sbjct 198 KVNLPENDFLSPDKLSLLEKLLPKRKEVEETDE 230
> At2g20550
Length=284
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 35/137 (25%), Positives = 63/137 (45%), Gaps = 5/137 (3%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L V++ G G+ + +G+E + G DL I + H TREG D+ + IS+
Sbjct 149 LTVDVKPGWKTGTKITFSEKGNE-QPGVIPADLVFIIDEKPHPVFTREGNDLVVTQKISV 207
Query 186 EDAVYGCSITVHTVDGDVELKIPALSRSGRKF--LIKGRGAKKIDDPSGARGDHVVTLRV 243
+A G ++ + T+DG L IP + ++ ++ G D +G+ + +
Sbjct 208 LEAFTGYTVNLTTLDGR-RLTIPVNTVIHPEYVEVVPNEGMPLQKD-QAKKGNLRIKFNI 265
Query 244 ALPETATAKDRELLKSL 260
P T T++ + LK L
Sbjct 266 KFPTTLTSEQKTGLKKL 282
> At5g01390
Length=335
Score = 42.7 bits (99), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 38/138 (27%), Positives = 65/138 (47%), Gaps = 7/138 (5%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L + I G +G+ + +G+E R G DL + + H R+G D+ + ISL
Sbjct 199 LTIEIKPGWKKGTKITFLEKGNEHR-GVIPSDLVFIVDEKPHPVFKRDGNDLVVMQKISL 257
Query 186 EDAVYGCSITVHTVDGDVELKIP---ALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLR 242
DA+ G + V T+DG L +P +S S + ++KG G DPS +G+ +
Sbjct 258 VDALTGYTAQVTTLDGRT-LTVPVNNVISPSYEE-VVKGEGMPIPKDPS-RKGNLRIRFI 314
Query 243 VALPETATAKDRELLKSL 260
+ P T + + +K +
Sbjct 315 IKFPSKLTTEQKSGIKRM 332
> At3g08910
Length=323
Score = 42.7 bits (99), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 62/136 (45%), Gaps = 3/136 (2%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L + I G +G+ + +G+E R G DL + + H R+G D+ + I L
Sbjct 186 LTIEIKPGWKKGTKITFPEKGNEQR-GIIPSDLVFIVDEKPHAVFKRDGNDLVMTQKIPL 244
Query 186 EDAVYGCSITVHTVDG-DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHVVTLRVA 244
+A+ G + V T+DG V + I + + ++KG G DPS +G+ + V
Sbjct 245 VEALTGYTAQVSTLDGRSVTVPINNVISPSYEEVVKGEGMPIPKDPS-KKGNLRIKFTVK 303
Query 245 LPETATAKDRELLKSL 260
P T + + +K +
Sbjct 304 FPSRLTTEQKSGIKRM 319
> 7295437
Length=334
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 35/130 (26%), Positives = 57/130 (43%), Gaps = 14/130 (10%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L++ + G G+ + EG N + D+ I + H REG D+ ISL
Sbjct 197 LRITVKPGWKAGTKITFPQEGDSAPN-KTPADIVFIIRDKPHSLFKREGIDLKYTAQISL 255
Query 186 EDAVYGCSITVHTVDG-------DVELKIPALSRSGRKFLIKGRGAKKIDDPSGARGDHV 238
+ A+ G ++V T+ G + E+ P +R I G G +PS RGD +
Sbjct 256 KQALCGALVSVPTLQGSRIQVNPNHEIIKPTTTRR-----INGLGLPVPKEPS-RRGDLI 309
Query 239 VTLRVALPET 248
V+ + P+T
Sbjct 310 VSFDIKFPDT 319
> At2g20560
Length=337
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 34/137 (24%), Positives = 65/137 (47%), Gaps = 5/137 (3%)
Query 126 LKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSEESISL 185
L +++ G +G+ + +G+E + G DL I + H TREG D+ + ISL
Sbjct 202 LTIDVKPGWKKGTKITFPEKGNE-QPGVIPADLVFIIDEKPHPVFTREGNDLIVTQKISL 260
Query 186 EDAVYGCSITVHTVDGDVELKIPALSRSGRKF--LIKGRGAKKIDDPSGARGDHVVTLRV 243
+A+ G ++ + T+DG L IP + ++ ++ G D + RG+ + +
Sbjct 261 VEALTGYTVNLTTLDGR-RLTIPVTNVVHPEYEEVVPKEGMPLQKDQT-KRGNLRIKFNI 318
Query 244 ALPETATAKDRELLKSL 260
P T++ + +K L
Sbjct 319 KFPTRLTSEQKTGVKKL 335
> Hs22064144
Length=141
Score = 40.8 bits (94), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 26/106 (24%), Positives = 47/106 (44%), Gaps = 4/106 (3%)
Query 156 GDLYISIGVESHDRLTREGADVCSEESISLEDAVYGCSITVHTVDGDVELKIPALSRSGR 215
D+ + + H R RE ++ I L A+ C++ V T+D D L IP
Sbjct 34 ADIIFIVKEKLHPRFRRENDNLFFVNPIPLGKALTCCTVEVRTLD-DRLLNIPINDIIHP 92
Query 216 KFL--IKGRGAKKIDDPSGARGDHVVTLRVALPETATAKDRELLKS 259
K+ + G G +DP+ +GD + + P T + +++L+
Sbjct 93 KYFKKVPGEGMPLPEDPT-KKGDLFIFFDIQFPTRLTPQKKQMLRQ 137
> YNL007c
Length=352
Score = 40.4 bits (93), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 32/131 (24%), Positives = 57/131 (43%), Gaps = 10/131 (7%)
Query 121 SKDVHLKVNIPAGVLEGSLLRVEGEGSEGRNGEARGDLYISIGVESHDRLTREGADVCSE 180
S+ + + + G G+ + + +G R L I +SH R+G D+
Sbjct 212 SEKTQIDIQLKPGWKAGTKITYKNQGDYNPQTGRRKTLQFVIQEKSHPNFKRDGDDLIYT 271
Query 181 ESISLEDAVYGCSITVHTVDGDVELKIPALSR-----SGRKFLIKGRGAKKIDDPSGARG 235
+S ++++ G S T+ T+DG +P LSR + G+G +PS RG
Sbjct 272 LPLSFKESLLGFSKTIQTIDGRT---LP-LSRVQPVQPSQTSTYPGQGMPTPKNPS-QRG 326
Query 236 DHVVTLRVALP 246
+ +V +V P
Sbjct 327 NLIVKYKVDYP 337
Lambda K H
0.314 0.131 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5702305406
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40