bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0527_orf3
Length=317
Score E
Sequences producing significant alignments: (Bits) Value
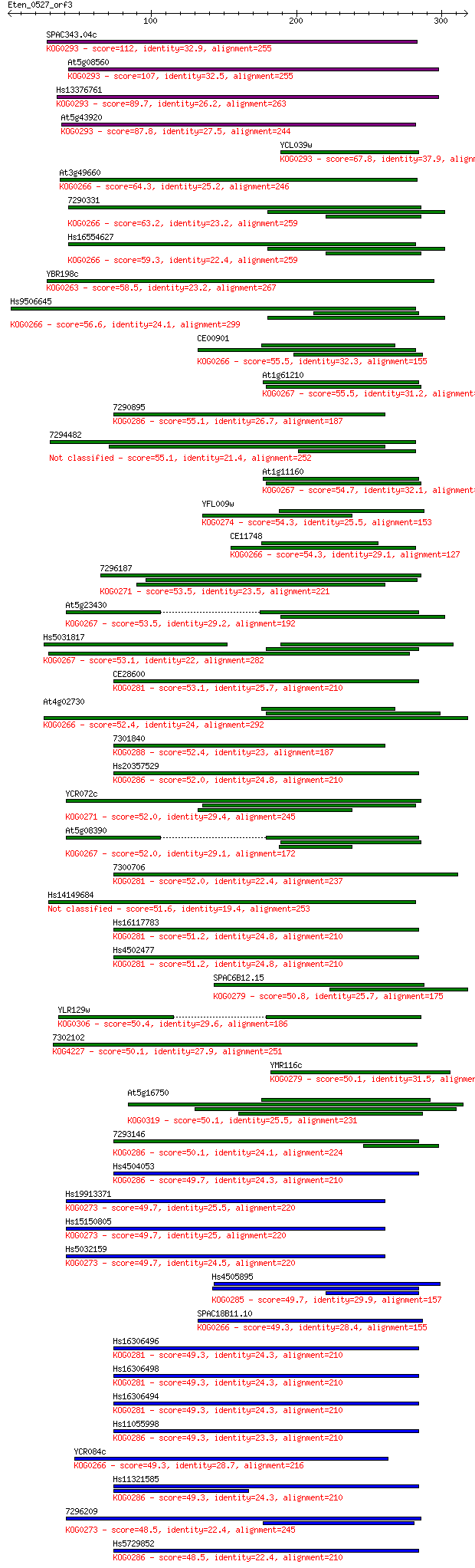
SPAC343.04c 112 1e-24
At5g08560 107 4e-23
Hs13376761 89.7 7e-18
At5g43920 87.8 3e-17
YCL039w 67.8 3e-11
At3g49660 64.3 3e-10
7290331 63.2 7e-10
Hs16554627 59.3 1e-08
YBR198c 58.5 2e-08
Hs9506645 56.6 6e-08
CE00901 55.5 1e-07
At1g61210 55.5 1e-07
7290895 55.1 2e-07
7294482 55.1 2e-07
At1g11160 54.7 2e-07
YFL009w 54.3 3e-07
CE11748 54.3 4e-07
7296187 53.5 6e-07
At5g23430 53.5 6e-07
Hs5031817 53.1 8e-07
CE28600 53.1 8e-07
At4g02730 52.4 1e-06
7301840 52.4 1e-06
Hs20357529 52.0 2e-06
YCR072c 52.0 2e-06
At5g08390 52.0 2e-06
7300706 52.0 2e-06
Hs14149684 51.6 2e-06
Hs16117783 51.2 2e-06
Hs4502477 51.2 3e-06
SPAC6B12.15 50.8 4e-06
YLR129w 50.4 5e-06
7302102 50.1 6e-06
YMR116c 50.1 7e-06
At5g16750 50.1 7e-06
7293146 50.1 7e-06
Hs4504053 49.7 7e-06
Hs19913371 49.7 8e-06
Hs15150805 49.7 9e-06
Hs5032159 49.7 9e-06
Hs4505895 49.7 9e-06
SPAC18B11.10 49.3 9e-06
Hs16306496 49.3 9e-06
Hs16306498 49.3 1e-05
Hs16306494 49.3 1e-05
Hs11055998 49.3 1e-05
YCR084c 49.3 1e-05
Hs11321585 49.3 1e-05
7296209 48.5 2e-05
Hs5729852 48.5 2e-05
> SPAC343.04c
Length=507
Score = 112 bits (280), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 84/261 (32%), Positives = 124/261 (47%), Gaps = 12/261 (4%)
Query 28 LWRVQRLA---SATAFLDWDSTGKLLAAGGEAAFIEAWE--EGRRIGDYCPHGGSIVALQ 82
L RV RL A++ W + L + + W+ G ++ DY HG S+
Sbjct 245 LKRVFRLIGHIDTVAYIRWSPDDRYLLSCSCDKSVILWDAFTGEKLRDY-KHGFSVSCCC 303
Query 83 WVPGSRRLISMATDRTMTLVSLEESSLTPTHVVE-YEWILPGRPQEGFLLPRGNSVMIFF 141
W+P I+ + D +T SL L V Y+ L + +++ G +I
Sbjct 304 WLPDGLSFITGSPDCHITHWSLNGEILYKWEDVNIYDMALTSDGTKLYIV--GFEQLINA 361
Query 142 ADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQ 201
D+ I ++ + T+E S + V + C SK L + P LWDL+E +IV+
Sbjct 362 EDKHIAIYSVETRECIKKISLQSKVTSICLSKDSKYALTNLE--PHTTFLWDLEENRIVR 419
Query 202 KYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWV 261
+Y GHK+G I FGG + V+SGSED +I IWHR G +L L GH VN VA+
Sbjct 420 QYMGHKLGNFLIGSCFGGKDDTFVLSGSEDDKIRIWHRESGKLLATLSGHVKCVNYVAY- 478
Query 262 NFCNPVCLLSAGDDGVLLLWS 282
N +P SAGDD + +WS
Sbjct 479 NPVDPYQFASAGDDNTVRIWS 499
> At5g08560
Length=589
Score = 107 bits (267), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 83/274 (30%), Positives = 126/274 (45%), Gaps = 40/274 (14%)
Query 43 WDSTGKLLAAGGEAAFIEAWE--EGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTMT 100
W + + G I W+ G + Y G S ++ W P + +I+ TDR++
Sbjct 327 WSPDDRQVLTCGAEEVIRRWDVDSGDCVHMYEKGGISPISCGWYPDGQGIIAGMTDRSIC 386
Query 101 LVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSV------------MIFFADRQIKL 148
+ W L GR +E + R V + D I L
Sbjct 387 M-----------------WDLDGREKECWKGQRTQKVSDIAMTDDGKWLVSVCKDSVISL 429
Query 149 FDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLD-ERKIVQKYKGHK 207
FD E E+D++ + S IL++ +RLW+++ + KIV +YKGHK
Sbjct 430 FDREATVERL-IEEEDMITSFSLSNDNKYILVNLLNQE--IRLWNIEGDPKIVSRYKGHK 486
Query 208 VGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPV 267
R IR FGG ++ + SGSED+Q+YIWHR G ++ +L GHA AVN V+W + N
Sbjct 487 RSRFIIRSCFGGYKQAFIASGSEDSQVYIWHRSTGKLIVELPGHAGAVNCVSW-SPTNLH 545
Query 268 CLLSAGDDGVLLLWSLPTTSNEER----VEPSSS 297
L SA DDG + +W L + + + V+ SSS
Sbjct 546 MLASASDDGTIRIWGLDRINQQNQKKKLVQGSSS 579
> Hs13376761
Length=367
Score = 89.7 bits (221), Expect = 7e-18, Method: Compositional matrix adjust.
Identities = 69/268 (25%), Positives = 115/268 (42%), Gaps = 16/268 (5%)
Query 35 ASATAFLDWDSTGKLLAAGGEAAFIEAWEEGRRIGDY-----CPHGGSIVALQWVPGSRR 89
A +++ W L A G E W + G+ H S+ ++ W P +R
Sbjct 107 AYGVSYIAWSPDDNYLVACGPDDCSELWLWNVQTGELRTKMSQSHEDSLTSVAWNPDGKR 166
Query 90 LISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADRQIKLF 149
++ L+ + L V R Q + L G +V+ ++I+ +
Sbjct 167 FVTGGQRGQFYQCDLDGNLLDSWEGV--------RVQCLWCLSDGKTVLASDTHQRIRGY 218
Query 150 DLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVG 209
+ + E +++ SK L++ AT + LWDL +R +V+KY+G G
Sbjct 219 NFEDLTDRNIVQEDHPIMSFTISKNGRLALLNVATQG--VHLWDLQDRVLVRKYQGVTQG 276
Query 210 RLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCL 269
I FGG E+ + SGSED ++YIWH+ + +L GH VN V+W N P +
Sbjct 277 FYTIHSCFGGHNEDFIASGSEDHKVYIWHKRSELPIAELTGHTRTVNCVSW-NPQIPSMM 335
Query 270 LSAGDDGVLLLWSLPTTSNEERVEPSSS 297
SA DDG + +W + + +E S
Sbjct 336 ASASDDGTVRIWGPAPFIDHQNIEEECS 363
> At5g43920
Length=523
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 67/249 (26%), Positives = 112/249 (44%), Gaps = 12/249 (4%)
Query 38 TAFLDWDSTGKLLAAGGEAAFIEAWEEG----RRIGDYCPHGGSIVALQWVPGSRRLISM 93
+F+ W L G A ++ W+ R G ++ + W P S RL+
Sbjct 272 VSFVSWSPDDTKLLTCGNAEVLKLWDVDTGVLRHTFGNNNTGFTVSSCAWFPDSTRLVCG 331
Query 94 ATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADRQIKLFDLVT 153
++D +V + + + + + + P G S++ F+D++I++ +L T
Sbjct 332 SSDPERGIVMWDTDG---NEIKAWRGTRIPKVVDLAVTPDGESMITVFSDKEIRILNLET 388
Query 154 KEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDL-DERKIVQKYKGHKVGRLA 212
K E + E+ + + + I + + LWDL E K K+ GH+ +
Sbjct 389 KVERVISEEQPITSLSISGD--GKFFIVNLSCQEI-HLWDLAGEWKQPLKFSGHRQSKYV 445
Query 213 IRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSA 272
IR FGG + SGSED+Q+YIW+ L+ L GH+ VN V+W N NP L SA
Sbjct 446 IRSCFGGLDSSFIASGSEDSQVYIWNLKNTKPLEVLSGHSMTVNCVSW-NPKNPRMLASA 504
Query 273 GDDGVLLLW 281
DD + +W
Sbjct 505 SDDQTIRIW 513
> YCL039w
Length=745
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 36/105 (34%), Positives = 53/105 (50%), Gaps = 11/105 (10%)
Query 189 LRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQL 248
L++WD E ++QKY G K IR F + LV+SGSED +IYIW R+ G ++ L
Sbjct 639 LQMWDYKENILIQKYFGQKQQHFIIRSCFAYGNK-LVMSGSEDGKIYIWDRIRGNLVSVL 697
Query 249 RGHAAAVN----------QVAWVNFCNPVCLLSAGDDGVLLLWSL 283
GH+ ++ V N + S GDDG + +W +
Sbjct 698 SGHSTVMSNSTKPMGKNCNVVASNPADKEMFASGGDDGKIKIWKI 742
> At3g49660
Length=317
Score = 64.3 bits (155), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 62/300 (20%), Positives = 117/300 (39%), Gaps = 66/300 (22%)
Query 37 ATAFLDWDSTGKLLAAGGEAAFIEAWEEG-------RRIGDYCPHGGSIVALQWVPGSRR 89
A + + + S G+LLA+ I + + ++ H I + + +R
Sbjct 26 AVSSVKFSSDGRLLASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSSDARF 85
Query 90 LISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLL---PRGNSVMIFFADRQI 146
++S + D+T+ L +E SL T L G F + P+ N ++ D +
Sbjct 86 IVSASDDKTLKLWDVETGSLIKT--------LIGHTNYAFCVNFNPQSNMIVSGSFDETV 137
Query 147 KLFDLVTKE-------------ETFWASEKDLVIAACPSKIC------------------ 175
+++D+ T + + + L++++ +C
Sbjct 138 RIWDVTTGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDSGTGHCVKTLIDDE 197
Query 176 -----------NQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREEL 224
N I T LRLW++ K ++ Y GH + I AF +
Sbjct 198 NPPVSFVRFSPNGKFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKR 257
Query 225 VISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPV-CLLSAGD-DGVLLLWS 282
++SGSED +++W +LQ+L GH V VA C+P L+++G D + +W+
Sbjct 258 IVSGSEDNCVHMWELNSKKLLQKLEGHTETVMNVA----CHPTENLIASGSLDKTVRIWT 313
> 7290331
Length=361
Score = 63.2 bits (152), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 57/248 (22%), Positives = 102/248 (41%), Gaps = 13/248 (5%)
Query 43 WDSTGKLLAAGGEAAFIEAWE--EGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTMT 100
W S +LL +G + ++ WE G+ + H + + P S ++S + D ++
Sbjct 122 WSSDSRLLVSGSDDKTLKVWELSTGKSLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVR 181
Query 101 LVSLEESSLTPTHVVEYEWILPGR--PQEGFLLPRGNSVMIFFA-DRQIKLFDLVTKEET 157
+ + T LP P R S+++ + D +++D + +
Sbjct 182 IWDVRTGKCLKT--------LPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCL 233
Query 158 FWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAF 217
+ D + N I AAT L+LWD + K ++ Y GHK + I F
Sbjct 234 KTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLKTYTGHKNEKYCIFANF 293
Query 218 GGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGV 277
+ ++SGSED +YIW+ ++Q+L+GH V A N + + +D
Sbjct 294 SVTGGKWIVSGSEDNMVYIWNLQSKEVVQKLQGHTDTVLCTACHPTENIIASAALENDKT 353
Query 278 LLLWSLPT 285
+ LW T
Sbjct 354 IKLWKSDT 361
Score = 40.4 bits (93), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 59/124 (47%), Gaps = 9/124 (7%)
Query 180 ISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHR 239
+++++ ++++W + K + GHK+G I L++SGS+D + +W
Sbjct 87 LASSSADKLIKIWGAYDGKFEKTISGHKLG---ISDVAWSSDSRLLVSGSDDKTLKVWEL 143
Query 240 VWGCMLQQLRGHAAAVNQVAWVNFCNPVC--LLSAGDDGVLLLWSLPTTSNEERVEPSSS 297
G L+ L+GH+ N V NF NP ++S D + +W + T + + S
Sbjct 144 STGKSLKTLKGHS---NYVFCCNF-NPQSNLIVSGSFDESVRIWDVRTGKCLKTLPAHSD 199
Query 298 PETA 301
P +A
Sbjct 200 PVSA 203
Score = 36.6 bits (83), Expect = 0.079, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 31/66 (46%), Gaps = 2/66 (3%)
Query 220 PREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLL 279
P E + S S D I IW G + + GH ++ VAW + + L+S DD L
Sbjct 82 PNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSS--DSRLLVSGSDDKTLK 139
Query 280 LWSLPT 285
+W L T
Sbjct 140 VWELST 145
> Hs16554627
Length=334
Score = 59.3 bits (142), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 54/244 (22%), Positives = 100/244 (40%), Gaps = 13/244 (5%)
Query 43 WDSTGKLLAAGGEAAFIEAWE--EGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTMT 100
W S LL + + ++ W+ G+ + H + + P S ++S + D ++
Sbjct 95 WSSDSNLLVSASDDKTLKIWDVSSGKCLKTLKGHSNYVFCCNFNPQSNLIVSGSFDESVR 154
Query 101 LVSLEESSLTPTHVVEYEWILPGR--PQEGFLLPRGNSVMIFFA-DRQIKLFDLVTKEET 157
+ ++ T LP P R S+++ + D +++D + +
Sbjct 155 IWDVKTGKCLKT--------LPAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCL 206
Query 158 FWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAF 217
+ D + N I AAT L+LWD + K ++ Y GHK + I F
Sbjct 207 KTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLWDYSKGKCLKTYTGHKNEKYCIFANF 266
Query 218 GGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGV 277
+ ++SGSED +YIW+ ++Q+L+GH V A N + + +D
Sbjct 267 SVTGGKWIVSGSEDNLVYIWNLQTKEIVQKLQGHTDVVISTACHPTENIIASAALENDKT 326
Query 278 LLLW 281
+ LW
Sbjct 327 IKLW 330
Score = 40.4 bits (93), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/124 (25%), Positives = 58/124 (46%), Gaps = 9/124 (7%)
Query 180 ISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHR 239
+++++ ++++W + K + GHK+G I L++S S+D + IW
Sbjct 60 LASSSADKLIKIWGAYDGKFEKTISGHKLG---ISDVAWSSDSNLLVSASDDKTLKIWDV 116
Query 240 VWGCMLQQLRGHAAAVNQVAWVNFCNPVC--LLSAGDDGVLLLWSLPTTSNEERVEPSSS 297
G L+ L+GH+ N V NF NP ++S D + +W + T + + S
Sbjct 117 SSGKCLKTLKGHS---NYVFCCNF-NPQSNLIVSGSFDESVRIWDVKTGKCLKTLPAHSD 172
Query 298 PETA 301
P +A
Sbjct 173 PVSA 176
Score = 38.1 bits (87), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query 220 PREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLL 279
P E + S S D I IW G + + GH ++ VAW + N L+SA DD L
Sbjct 55 PNGEWLASSSADKLIKIWGAYDGKFEKTISGHKLGISDVAWSSDSN--LLVSASDDKTLK 112
Query 280 LWSLPT 285
+W + +
Sbjct 113 IWDVSS 118
> YBR198c
Length=798
Score = 58.5 bits (140), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 62/290 (21%), Positives = 115/290 (39%), Gaps = 37/290 (12%)
Query 28 LWRVQRLASATAFLDWDSTGKLLAAGGEAAFIEAW-------------------EEGRRI 68
++ Q + LD+ ++ AAG + ++I+ W +E
Sbjct 459 MYTFQNTNKDMSCLDFSDDCRIAAAGFQDSYIKIWSLDGSSLNNPNIALNNNDKDEDPTC 518
Query 69 GDYCPHGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQ-E 127
H G++ + + P ++ L+S + D+T+ L S++ TH + P +
Sbjct 519 KTLVGHSGTVYSTSFSPDNKYLLSGSEDKTVRLWSMD------THTALVSYKGHNHPVWD 572
Query 128 GFLLPRGNSVMIFFADRQIKLF--DLVTKEETFWASEKDLVIAACPSKICNQILISAATT 185
P G+ D+ +L+ D + F D+ C S N + ++
Sbjct 573 VSFSPLGHYFATASHDQTARLWSCDHIYPLRIFAGHLNDV---DCVSFHPNGCYVFTGSS 629
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
R+WD+ V+ + GH ++I P + +GSED I +W G L
Sbjct 630 DKTCRMWDVSTGDSVRLFLGHTAPVISIAVC---PDGRWLSTGSEDGIINVWDIGTGKRL 686
Query 246 QQLRGHAA-AVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEP 294
+Q+RGH A+ +++ N L+S G D + +W L + E EP
Sbjct 687 KQMRGHGKNAIYSLSYSKEGN--VLISGGADHTVRVWDLKKATTEPSAEP 734
> Hs9506645
Length=330
Score = 56.6 bits (135), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 68/330 (20%), Positives = 119/330 (36%), Gaps = 67/330 (20%)
Query 3 LLMRLEQTQARPILESGNVIGTCRLLWRVQRLASATAFLDWDSTGKLLAAGGEAAFIEAW 62
L + Q++ + E+ N C L+ + ++S + + G+ LA+ I W
Sbjct 13 LALSSSANQSKEVPENPNYALKCTLVGHTEAVSS----VKFSPNGEWLASSSADRLIIIW 68
Query 63 --EEGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWI 120
+G+ H I + W S RL+S + D+T+ L + T
Sbjct 69 GAYDGKYEKTLYGHNLEISDVAWSSDSSRLVSASDDKTLKLWDVRSGKCLKT-------- 120
Query 121 LPGRPQEGFLL---PRGNSVMIFFADRQIKLFDLVTKE-------------ETFWASEKD 164
L G F P N ++ D +K++++ T + +
Sbjct 121 LKGHSNYVFCCNFNPPSNLIISGSFDETVKIWEVKTGKCLKTLSAHSDPVSAVHFNCSGS 180
Query 165 LVIAACPSKIC-----------------------------NQILISAATTPPVLRLWDLD 195
L+++ +C N I AT L+LWD
Sbjct 181 LIVSGSYDGLCRIWDAASGQCLKTLVDDDNPPVSFVKFSPNGKYILTATLDNTLKLWDYS 240
Query 196 ERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAV 255
+ ++ Y G K + I F + ++SGSED +YIW+ ++Q+L+GH V
Sbjct 241 RGRCLKTYTGQKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQKLQGHTDVV 300
Query 256 NQVAWVNFCNPVCLLSAG----DDGVLLLW 281
A C+P L A +D + LW
Sbjct 301 ISAA----CHPTENLIASAALENDKTIKLW 326
Score = 35.4 bits (80), Expect = 0.17, Method: Compositional matrix adjust.
Identities = 24/72 (33%), Positives = 33/72 (45%), Gaps = 2/72 (2%)
Query 212 AIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLS 271
A+ P E + S S D I IW G + L GH ++ VAW + + L+S
Sbjct 43 AVSSVKFSPNGEWLASSSADRLIIIWGAYDGKYEKTLYGHNLEISDVAWSS--DSSRLVS 100
Query 272 AGDDGVLLLWSL 283
A DD L LW +
Sbjct 101 ASDDKTLKLWDV 112
Score = 33.5 bits (75), Expect = 0.67, Method: Compositional matrix adjust.
Identities = 28/123 (22%), Positives = 54/123 (43%), Gaps = 7/123 (5%)
Query 180 ISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHR 239
+++++ ++ +W + K + GH L I ++S S+D + +W
Sbjct 56 LASSSADRLIIIWGAYDGKYEKTLYGH---NLEISDVAWSSDSSRLVSASDDKTLKLWDV 112
Query 240 VWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLWSLPTTSNEERVEPSSSP 298
G L+ L+GH+ N V NF P L+ +G D + +W + T + + S P
Sbjct 113 RSGKCLKTLKGHS---NYVFCCNFNPPSNLIISGSFDETVKIWEVKTGKCLKTLSAHSDP 169
Query 299 ETA 301
+A
Sbjct 170 VSA 172
> CE00901
Length=376
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Query 176 NQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIY 235
N I A+ L+LWD + K +++Y GH+ + I F + +ISGSED +IY
Sbjct 267 NGKYILASNLDSTLKLWDFSKGKTLKQYTGHENSKYCIFANFSVTGGKWIISGSEDCKIY 326
Query 236 IWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPV 267
IW+ ++Q L GH Q + C+PV
Sbjct 327 IWNLQTREIVQCLEGHT----QPVLASDCHPV 354
Score = 38.5 bits (88), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 37/164 (22%), Positives = 69/164 (42%), Gaps = 34/164 (20%)
Query 132 PRGNSVMIFFADRQIKL--FDLVTKEETF-----------WASEKDLVIAACPSKICNQI 178
P G + AD+ +K+ D + E T W+S+ V++A K
Sbjct 97 PCGKYLGTSSADKTVKIWNMDHMICERTLTGHKLGVNDIAWSSDSRCVVSASDDK----- 151
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWH 238
L+++++ ++ + KGH P+ LV+SGS D + IW
Sbjct 152 ---------TLKIFEIVTSRMTKTLKGHNNYVFCCN---FNPQSSLVVSGSFDESVRIWD 199
Query 239 RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLW 281
G ++ L H+ + V+ V+F L+++G DG++ +W
Sbjct 200 VKTGMCIKTLPAHS---DPVSAVSFNRDGSLIASGSYDGLVRIW 240
Score = 35.8 bits (81), Expect = 0.14, Method: Compositional matrix adjust.
Identities = 24/89 (26%), Positives = 43/89 (48%), Gaps = 5/89 (5%)
Query 198 KIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQ 257
K++ +GH +I A P + + + S D + IW+ + L GH VN
Sbjct 78 KLMCTLEGH---TKSISSAKFSPCGKYLGTSSADKTVKIWNMDHMICERTLTGHKLGVND 134
Query 258 VAWVNFCNPVCLLSAGDDGVLLLWSLPTT 286
+AW + + C++SA DD L ++ + T+
Sbjct 135 IAWSS--DSRCVVSASDDKTLKIFEIVTS 161
> At1g61210
Length=282
Score = 55.5 bits (132), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/108 (30%), Positives = 60/108 (55%), Gaps = 7/108 (6%)
Query 177 QILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYI 236
++L+ A + V++LWD++E K+V+ + GH+ A+ P E + SGS DA + I
Sbjct 100 EVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFH---PFGEFLASGSSDANLKI 156
Query 237 WH-RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
W R GC +Q +GH+ ++ + + + ++S G D V+ +W L
Sbjct 157 WDIRKKGC-IQTYKGHSRGISTIRFTP--DGRWVVSGGLDNVVKVWDL 201
Score = 45.1 bits (105), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 50/108 (46%), Gaps = 7/108 (6%)
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWH 238
+++ ++ L++WD+ ++ +Q YKGH G IR P V+SG D + +W
Sbjct 144 FLASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFT---PDGRWVVSGGLDNVVKVWD 200
Query 239 RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLWSLPT 285
G +L + + H + ++F LL+ G D + W L T
Sbjct 201 LTAGKLLHEFKFHEGPIRS---LDFHPLEFLLATGSADRTVKFWDLET 245
> 7290895
Length=358
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 50/201 (24%), Positives = 82/201 (40%), Gaps = 31/201 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTL-----VSLEESSLTPTHVVEYEWILPGRPQEG 128
H ++ W P R +IS + D + + + E + PT WI+
Sbjct 69 HQAKVLCTDWSPDKRHIISSSQDGRLIIWDAFTTNKEHAVTMPT-----TWIMACA---- 119
Query 129 FLLPRGNSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKIC-------NQILIS 181
P GN V D ++ ++ + + EE A++K V C QIL
Sbjct 120 -YAPSGNFVACGGLDNKVTVYPITSDEEM--AAKKRTVGTHTSYMSCCIYPNSDQQILTG 176
Query 182 AATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPRE--ELVISGSEDAQIYIWHR 239
+ + LWD++ +++Q + GH +AI A P E +SGS D +IW
Sbjct 177 SGDS--TCALWDVESGQLLQSFHGHSGDVMAIDLA---PNETGNTFVSGSCDRMAFIWDM 231
Query 240 VWGCMLQQLRGHAAAVNQVAW 260
G ++Q GH + VN V +
Sbjct 232 RSGHVVQSFEGHQSDVNSVKF 252
> 7294482
Length=391
Score = 55.1 bits (131), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 54/262 (20%), Positives = 113/262 (43%), Gaps = 26/262 (9%)
Query 30 RVQRLASATAFLD---WDSTGKLLAAGGEAAFIEAWEEGRR--IGDYCPHGGSIVALQWV 84
R R AS +A ++ W G L+A+ G ++ WE R G++ H ++ ++ +
Sbjct 52 RCIRFASHSAPVNGVAWSPKGNLVASAGHDRTVKIWEPKLRGVSGEFVAHSKAVRSVDFD 111
Query 85 PGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADR 144
+++ + D++ + + + + W+ + P G V D+
Sbjct 112 STGHLMLTASDDKSAKIWRVARRQFVSSFAQQNNWVRSAK-----FSPNGKLVATASDDK 166
Query 145 QIKLFDLVTKEETFWASEKDLVIAACPSKICNQ---ILISAATTPPVLRLWDLDERKIVQ 201
++++D+ + E +E+ A P ++ +++ A ++++D+ +++Q
Sbjct 167 SVRIYDVDSGECVRTFTEE----RAAPRQLAWHPWGNMLAVALGCNRIKIFDVSGSQLLQ 222
Query 202 KYKGHK--VGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVA 259
Y H V +A P+ ++SGS+D I I + G + L GH AVN VA
Sbjct 223 LYVVHSAPVNDVAFHPS-----GHFLLSGSDDRTIRILDLLEGRPIYTLTGHTDAVNAVA 277
Query 260 WVNFCNPVCLLSAGDDGVLLLW 281
+ + + G D LL+W
Sbjct 278 FSRDGDKFA--TGGSDRQLLVW 297
Score = 35.8 bits (81), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 41/193 (21%), Positives = 80/193 (41%), Gaps = 15/193 (7%)
Query 71 YCPHGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFL 130
+ H G I L++ P ++ + +TD T+ L +L +++ + P G
Sbjct 14 FTGHSGGITQLRFGPDGAQIATSSTDSTVILWNLNQAARC------IRFASHSAPVNGVA 67
Query 131 -LPRGNSVMIFFADRQIKLFD--LVTKEETFWASEKDLVIAACPSKICNQILISAATTPP 187
P+GN V DR +K+++ L F A K + + ++++A+
Sbjct 68 WSPKGNLVASAGHDRTVKIWEPKLRGVSGEFVAHSK--AVRSVDFDSTGHLMLTASDDKS 125
Query 188 VLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQ 247
++W + R+ V + +R A P +LV + S+D + I+ G ++
Sbjct 126 A-KIWRVARRQFVSSFAQQNNW---VRSAKFSPNGKLVATASDDKSVRIYDVDSGECVRT 181
Query 248 LRGHAAAVNQVAW 260
AA Q+AW
Sbjct 182 FTEERAAPRQLAW 194
Score = 33.9 bits (76), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 36/81 (44%), Gaps = 5/81 (6%)
Query 201 QKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAW 260
+ + GH G +R GP + + S D+ + +W+ + H+A VN VAW
Sbjct 12 RHFTGHSGGITQLRF---GPDGAQIATSSTDSTVILWNLNQAARCIRFASHSAPVNGVAW 68
Query 261 VNFCNPVCLLSAGDDGVLLLW 281
N V SAG D + +W
Sbjct 69 SPKGNLVA--SAGHDRTVKIW 87
> At1g11160
Length=974
Score = 54.7 bits (130), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 34/111 (30%), Positives = 60/111 (54%), Gaps = 13/111 (11%)
Query 177 QILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIR-PAFGGPREELVISGSEDAQIY 235
++L+ A + V++LWDL+E K+V+ + GH+ A+ FG E + SGS D +
Sbjct 19 EVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFHPFG----EFLASGSSDTNLR 74
Query 236 IWH-RVWGCMLQQLRGHAAAVNQVAWVNFCNPVC--LLSAGDDGVLLLWSL 283
+W R GC +Q +GH ++ + + +P ++S G D V+ +W L
Sbjct 75 VWDTRKKGC-IQTYKGHTRGISTIEF----SPDGRWVVSGGLDNVVKVWDL 120
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 28/108 (25%), Positives = 49/108 (45%), Gaps = 7/108 (6%)
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWH 238
+++ ++ LR+WD ++ +Q YKGH G I + P V+SG D + +W
Sbjct 63 FLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFS---PDGRWVVSGGLDNVVKVWD 119
Query 239 RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLWSLPT 285
G +L + + H + ++F LL+ G D + W L T
Sbjct 120 LTAGKLLHEFKCHEGPIRS---LDFHPLEFLLATGSADRTVKFWDLET 164
> YFL009w
Length=779
Score = 54.3 bits (129), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 28/100 (28%), Positives = 50/100 (50%), Gaps = 4/100 (4%)
Query 188 VLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQ 247
++R++D +K + + GH G A++ A GG +++SGS D + +W GC
Sbjct 403 MIRVYDSINKKFLLQLSGHDGGVWALKYAHGG----ILVSGSTDRTVRVWDIKKGCCTHV 458
Query 248 LRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTS 287
+GH + V + V + N +++ D L +W LP S
Sbjct 459 FKGHNSTVRCLDIVEYKNIKYIVTGSRDNTLHVWKLPKES 498
Score = 31.6 bits (70), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 21/103 (20%), Positives = 47/103 (45%), Gaps = 3/103 (2%)
Query 135 NSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDL 194
N V+ D+ I+++D + K+ S D + A K + ++ + +T +R+WD+
Sbjct 393 NYVITGADDKMIRVYDSINKKFLLQLSGHDGGVWAL--KYAHGGILVSGSTDRTVRVWDI 450
Query 195 DERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIW 237
+ +KGH + + +++GS D +++W
Sbjct 451 KKGCCTHVFKGHN-STVRCLDIVEYKNIKYIVTGSRDNTLHVW 492
> CE11748
Length=395
Score = 54.3 bits (129), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 44/80 (55%), Gaps = 0/80 (0%)
Query 176 NQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIY 235
N I A+ L+LWD + +++++Y GH+ + + F + ++SGSED ++Y
Sbjct 286 NGKYILASNLNNTLKLWDYQKLRVLKEYTGHENSKYCVAANFSVTGGKWIVSGSEDHKVY 345
Query 236 IWHRVWGCMLQQLRGHAAAV 255
IW+ +LQ L GH AV
Sbjct 346 IWNLQTREILQTLDGHNTAV 365
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 30/127 (23%), Positives = 52/127 (40%), Gaps = 19/127 (14%)
Query 155 EETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIR 214
E W+S+ L+++ K+ ++++D+ + V+ KGH
Sbjct 152 NEFSWSSDSKLIVSCSDDKL--------------VKVFDVSSGRCVKTLKGHT--NYVFC 195
Query 215 PAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGD 274
F P L+ SGS D I IW G + + GH V+ V + + L S
Sbjct 196 CCFN-PSGTLIASGSFDETIRIWCARNGNTIFSIPGHEDPVSSVCFNR--DGAYLASGSY 252
Query 275 DGVLLLW 281
DG++ +W
Sbjct 253 DGIVRIW 259
> 7296187
Length=481
Score = 53.5 bits (127), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 52/221 (23%), Positives = 91/221 (41%), Gaps = 13/221 (5%)
Query 65 GRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGR 124
G+ + + H ++ A++W G+ + + + DRT+ + + L T W+
Sbjct 234 GQCLMNIAGHTNAVTAVRW-GGAGLIYTSSKDRTVKMWRAADGILCRTFSGHAHWVNNIA 292
Query 125 PQEGFLLPRGNSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAAT 184
++L G + DR L T+E S A CP ++ + + S
Sbjct 293 LSTDYVLRTGPFHPV--KDRSKSHLSLSTEE--LQESALKRYQAVCPDEVESLVSCSDDN 348
Query 185 TPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCM 244
T L LW ++ K V++ GH+ + P +L+ S S D + +W G
Sbjct 349 T---LYLWRNNQNKCVERMTGHQN---VVNDVKYSPDVKLIASASFDKSVRLWRASDGQY 402
Query 245 LQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPT 285
+ RGH AV VAW + ++S D L +WS+ T
Sbjct 403 MATFRGHVQAVYTVAWS--ADSRLIVSGSKDSTLKVWSVQT 441
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/187 (22%), Positives = 81/187 (43%), Gaps = 15/187 (8%)
Query 96 DRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADRQIKLFDLVTKE 155
DR+ + +SL L + + Y+ + P E + ++ + + + Q K + +T
Sbjct 309 DRSKSHLSLSTEELQESALKRYQAVCPDEV-ESLVSCSDDNTLYLWRNNQNKCVERMTGH 367
Query 156 ETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRP 215
+ + P + LI++A+ +RLW + + + ++GH A+
Sbjct 368 QNVVND-----VKYSP----DVKLIASASFDKSVRLWRASDGQYMATFRGHV---QAVYT 415
Query 216 AFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDD 275
L++SGS+D+ + +W + Q+L GHA V V W + V S G D
Sbjct 416 VAWSADSRLIVSGSKDSTLKVWSVQTKKLAQELPGHADEVFGVDWAPDGSRVA--SGGKD 473
Query 276 GVLLLWS 282
V+ LW+
Sbjct 474 KVIKLWA 480
Score = 33.9 bits (76), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 42/178 (23%), Positives = 70/178 (39%), Gaps = 9/178 (5%)
Query 90 LISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLL---PRGNSVMIFFADRQI 146
L S+ T+ + +V ++ V +PG + L P G + D +
Sbjct 77 LASVDTENVIDIVYQPQAVFKVRPVTRCTSSMPGHAEAVVSLNFSPDGAHLASGSGDTTV 136
Query 147 KLFDLVTKEETFWAS-EKDLVIAACPSKICNQILISAATTPPVLRLWDLDE-RKIVQKYK 204
+L+DL T+ F + K V+ C S + +++ + +WD + ++ +
Sbjct 137 RLWDLNTETPHFTCTGHKQWVL--CVSWAPDGKRLASGCKAGSIIIWDPETGQQKGRPLS 194
Query 205 GHK--VGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAW 260
GHK + LA P P + S S D IW G L + GH AV V W
Sbjct 195 GHKKHINCLAWEPYHRDPECRKLASASGDGDCRIWDVKLGQCLMNIAGHTNAVTAVRW 252
> At5g23430
Length=922
Score = 53.5 bits (127), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 32/110 (29%), Positives = 57/110 (51%), Gaps = 7/110 (6%)
Query 175 CNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQI 234
+++L++A ++LWDL+E KIV+ GH+ +++ P E SGS D +
Sbjct 69 ASEVLVAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFH---PFGEFFASGSLDTNL 125
Query 235 YIWH-RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
IW R GC + +GH VN + + + ++S G+D ++ +W L
Sbjct 126 KIWDIRKKGC-IHTYKGHTRGVNVLRFT--PDGRWVVSGGEDNIVKVWDL 172
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 33/114 (28%), Positives = 52/114 (45%), Gaps = 11/114 (9%)
Query 189 LRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQL 248
L++WD+ ++ + YKGH G +R P V+SG ED + +W G +L +
Sbjct 125 LKIWDIRKKGCIHTYKGHTRGVNVLRFT---PDGRWVVSGGEDNIVKVWDLTAGKLLTEF 181
Query 249 RGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLWSLPTTSNEERVEPSSSPETA 301
+ H Q+ ++F LL+ G D + W L T + S PETA
Sbjct 182 KSHEG---QIQSLDFHPHEFLLATGSADRTVKFWDLETF----ELIGSGGPETA 228
Score = 32.0 bits (71), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 18/67 (26%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 41 LDWDSTGKLLAAGGEAAFIEAWE--EGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRT 98
L + G+ + +GGE ++ W+ G+ + ++ H G I +L + P L + + DRT
Sbjct 149 LRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHEGQIQSLDFHPHEFLLATGSADRT 208
Query 99 MTLVSLE 105
+ LE
Sbjct 209 VKFWDLE 215
> Hs5031817
Length=655
Score = 53.1 bits (126), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 33/119 (27%), Positives = 52/119 (43%), Gaps = 5/119 (4%)
Query 189 LRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQL 248
++LWD+ + V +Y+GH A+R P + + S ++D + +W G M+ +
Sbjct 129 IKLWDIRRKGCVFRYRGHS---QAVRCLRFSPDGKWLASAADDHTVKLWDLTAGKMMSEF 185
Query 249 RGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSSPETATLSPPD 307
GH VN V + N L S DG + W L R+E P + L PD
Sbjct 186 PGHTGPVNVVEF--HPNEYLLASGSSDGTIRFWDLEKFQVVSRIEGEPGPVRSVLFNPD 242
Score = 46.6 bits (109), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 36/106 (33%), Positives = 53/106 (50%), Gaps = 7/106 (6%)
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWH 238
LI A + +R+WDL+ KI++ G K ++ P E V SGS+D I +W
Sbjct 77 LIVAGSQSGSIRVWDLEAAKILRTLMGLKANICSLD---FHPYGEFVASGSQDTNIKLWD 133
Query 239 -RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
R GC+ + RGH+ AV + + + L SA DD + LW L
Sbjct 134 IRRKGCVF-RYRGHSQAVRCLRFSP--DGKWLASAADDHTVKLWDL 176
Score = 37.4 bits (85), Expect = 0.046, Method: Compositional matrix adjust.
Identities = 35/136 (25%), Positives = 61/136 (44%), Gaps = 23/136 (16%)
Query 26 RLLWRVQRLASATAFLDWDSTGKLLAAGGEAAFIEAWEEGRR--IGDYCPHGGSIVALQW 83
++L + L + LD+ G+ +A+G + I+ W+ R+ + Y H ++ L++
Sbjct 96 KILRTLMGLKANICSLDFHPYGEFVASGSQDTNIKLWDIRRKGCVFRYRGHSQAVRCLRF 155
Query 84 VPGSRRLISMATDRTMTLVSLEESSLT--------PTHVVEYEWILPGRPQEGFLLPRGN 135
P + L S A D T+ L L + P +VVE+ P E +LL G+
Sbjct 156 SPDGKWLASAADDHTVKLWDLTAGKMMSEFPGHTGPVNVVEFH------PNE-YLLASGS 208
Query 136 SVMIFFADRQIKLFDL 151
S D I+ +DL
Sbjct 209 S------DGTIRFWDL 218
Score = 36.6 bits (83), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 56/257 (21%), Positives = 102/257 (39%), Gaps = 21/257 (8%)
Query 29 WRVQRL----ASATAFLDWDSTGKLLAAGGEAAFIEAWEEGRR--IGDYCPHGGSIVALQ 82
W++Q + ++ ++ + ++G+LLA GG+ + W + I H + +++
Sbjct 11 WKLQEIVAHASNVSSLVLGKASGRLLATGGDDCRVNLWSINKPNCIMSLTGHTSPVESVR 70
Query 83 WVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFA 142
+++ + ++ + LE + + T + L P G V
Sbjct 71 LNTPEELIVAGSQSGSIRVWDLEAAKILRTLMG-----LKANICSLDFHPYGEFVASGSQ 125
Query 143 DRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQK 202
D IKL+D+ K F + + L SAA V +LWDL K++ +
Sbjct 126 DTNIKLWDIRRKGCVFRYRGHSQAVRCLRFSPDGKWLASAADDHTV-KLWDLTAGKMMSE 184
Query 203 YKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVN 262
+ GH G + + P E L+ SGS D I W ++ ++ G V V +
Sbjct 185 FPGH-TGPVNV--VEFHPNEYLLASGSSDGTIRFWDLEKFQVVSRIEGEPGPVRSVLF-- 239
Query 263 FCNP--VCLLSAGDDGV 277
NP CL S D +
Sbjct 240 --NPDGCCLYSGCQDSL 254
> CE28600
Length=665
Score = 53.1 bits (126), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 54/218 (24%), Positives = 95/218 (43%), Gaps = 34/218 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ +R +IS ++D T+ + +E T + E +L R G ++
Sbjct 261 HTGSVLCLQY--DNRVIISGSSDATVRVWDVETGECIKTLIHHCEAVLHLRFANGIMV-- 316
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVI--------AACPSKICNQILISAATT 185
DR I ++D+V S +D+ I AA + I +A+
Sbjct 317 -----TCSKDRSIAVWDMV--------SPRDITIRRVLVGHRAAVNVVDFDDRYIVSASG 363
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
+++W +D + V+ GH+ G ++ R LV+SGS D I +W G L
Sbjct 364 DRTIKVWSMDTLEFVRTLAGHRRGIACLQY-----RGRLVVSGSSDNTIRLWDIHSGVCL 418
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+ L GH V + + + ++S DG + +W L
Sbjct 419 RVLEGHEELVRCIRF----DEKRIVSGAYDGKIKVWDL 452
> At4g02730
Length=333
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 31/92 (33%), Positives = 44/92 (47%), Gaps = 4/92 (4%)
Query 176 NQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIY 235
N I AT L+L + K ++ Y GH I AF + ++SGSED +Y
Sbjct 224 NGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCITSAFSVTNGKYIVSGSEDNCVY 283
Query 236 IWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPV 267
+W +LQ+L GH AV V+ C+PV
Sbjct 284 LWDLQARNILQRLEGHTDAVISVS----CHPV 311
Score = 40.4 bits (93), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 33/123 (26%), Positives = 53/123 (43%), Gaps = 10/123 (8%)
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIW- 237
L+++A+ + LW ++ +Y+GH G I S S+D + IW
Sbjct 57 LLASASVDKTMILWSATNYSLIHRYEGHSSG---ISDLAWSSDSHYTCSASDDCTLRIWD 113
Query 238 -HRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLWSLPTTSNEERVEPS 295
+ C L+ LRGH N V VNF P L+ +G D + +W + T ++
Sbjct 114 ARSPYEC-LKVLRGH---TNFVFCVNFNPPSNLIVSGSFDETIRIWEVKTGKCVRMIKAH 169
Query 296 SSP 298
S P
Sbjct 170 SMP 172
Score = 35.0 bits (79), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 55/300 (18%), Positives = 112/300 (37%), Gaps = 20/300 (6%)
Query 26 RLLWRVQRLASATAFLDWDSTGKLLAAGGEAAFIEAWEEGRR--IGDYCPHGGSIVALQW 83
R L ++ +A + + + + G LLA+ + W I Y H I L W
Sbjct 34 RHLKTLEGHTAAISCVKFSNDGNLLASASVDKTMILWSATNYSLIHRYEGHSSGISDLAW 93
Query 84 VPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLL-----PRGNSVM 138
S S + D T+ + YE + R F+ P N ++
Sbjct 94 SSDSHYTCSASDDCTLRIWDARS---------PYECLKVLRGHTNFVFCVNFNPPSNLIV 144
Query 139 IFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERK 198
D I+++++ T + + I++ +++SA+ ++WD E
Sbjct 145 SGSFDETIRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSLIVSASHDGSC-KIWDAKEGT 203
Query 199 IVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQV 258
++ K A+ A P + ++ + D+ + + + G L+ GH V +
Sbjct 204 CLKTLIDDK--SPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTGHTNKVFCI 261
Query 259 -AWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSSPETATLSPPDQNTPDASSDQ 317
+ + N ++S +D + LW L + +R+E + + P QN +S +
Sbjct 262 TSAFSVTNGKYIVSGSEDNCVYLWDLQARNILQRLEGHTDAVISVSCHPVQNEISSSGNH 321
> 7301840
Length=220
Score = 52.4 bits (124), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 43/191 (22%), Positives = 88/191 (46%), Gaps = 15/191 (7%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H G ++A ++V ++++ + DRT+ + L + T G +
Sbjct 19 HSGKVMAAKYVQEPIKVVTGSHDRTLKIWDLRSIACIETK-------FAGSSCNDLVTTD 71
Query 134 --GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRL 191
G++++ D++I+ +D+ T+++ + + SK CN ++ S ++L
Sbjct 72 SLGSTIISGHYDKKIRFWDIRTEKQADDVLMPAKITSLDLSKDCNYLICSVR--DDTIKL 129
Query 192 WDLDERKIVQKYKGH--KVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLR 249
DL + +++ + K+ R +F ++ GS D IYIW+ V G + L+
Sbjct 130 LDLRKNQVISTFTNEHFKISCDFARASFNSSGLKIAC-GSADGAIYIWN-VNGFLEATLK 187
Query 250 GHAAAVNQVAW 260
GH+ AVN V+W
Sbjct 188 GHSTAVNAVSW 198
> Hs20357529
Length=340
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/216 (24%), Positives = 84/216 (38%), Gaps = 20/216 (9%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVV--EYEWILPGRPQEGFLL 131
H I A+ W SR L+S + D L+ + + H + W++
Sbjct 54 HLAKIYAMHWGTDSRLLVSASQDGK--LIIWDSYTTNKVHAIPLRSSWVMTCA-----YA 106
Query 132 PRGNSVMIFFADRQIKLFDLVTKEETFWASEK----DLVIAACPSKICNQILISAATTPP 187
P GN V D ++ L T+E S + ++ C NQI+ S+ T
Sbjct 107 PSGNFVACGGLDNICSIYSLKTREGNVRVSRELPGHTGYLSCCRFLDDNQIITSSGDTTC 166
Query 188 VLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQ 247
L WD++ + + GH +++ A P +SG+ DA I +W Q
Sbjct 167 AL--WDIETGQQTVGFAGHSGDVMSLSLA---PDGRTFVSGACDASIKLWDVRDSMCRQT 221
Query 248 LRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
GH + +N VA+ F N + DD L+ L
Sbjct 222 FIGHESDINAVAF--FPNGYAFTTGSDDATCRLFDL 255
> YCR072c
Length=515
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 72/297 (24%), Positives = 119/297 (40%), Gaps = 65/297 (21%)
Query 41 LDWDSTGKLLAAGGEAAFIEAWE--EGRRIGDYC-PHGGSIVALQW------VPGSR-RL 90
+ W G+++A G I W+ G+ +GD H I +L W PGS+ RL
Sbjct 192 VSWSPDGEVIATGSMDNTIRLWDPKSGQCLGDALRGHSKWITSLSWEPIHLVKPGSKPRL 251
Query 91 ISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFA--DRQIKL 148
S + D T+ + T + V +Y + G + G +++ DR +++
Sbjct 252 ASSSKDGTIKIWD------TVSRVCQY--TMSGHTNSVSCVKWGGQGLLYSGSHDRTVRV 303
Query 149 FDL--------VTKEETFW------ASEKDLVIAAC------PS--------------KI 174
+D+ + K W +++ L I A PS KI
Sbjct 304 WDINSQGRCINILKSHAHWVNHLSLSTDYALRIGAFDHTGKKPSTPEEAQKKALENYEKI 363
Query 175 CNQI-----LISAATTPPVLRLWD-LDERKIVQKYKGHKVGRLAIRPAFGGPREELVISG 228
C + ++ A+ + LW+ L K + + GH+ +L AF P ++S
Sbjct 364 CKKNGNSEEMMVTASDDYTMFLWNPLKSTKPIARMTGHQ--KLVNHVAFS-PDGRYIVSA 420
Query 229 SEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPT 285
S D I +W G + RGH A+V QVAW + C L+S D L +W + T
Sbjct 421 SFDNSIKLWDGRDGKFISTFRGHVASVYQVAWSSDCR--LLVSCSKDTTLKVWDVRT 475
Score = 43.1 bits (100), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 33/153 (21%), Positives = 67/153 (43%), Gaps = 10/153 (6%)
Query 135 NSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDL 194
+ + + + R + VT+ + A ++ + + + +++ A R+WD
Sbjct 115 DQITLLYTPRAVFKVKPVTRSSSAIAGHGSTILCSAFAPHTSSRMVTGAGDN-TARIWDC 173
Query 195 DERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWG-CMLQQLRGHAA 253
D + + KGH L + + P E++ +GS D I +W G C+ LRGH+
Sbjct 174 DTQTPMHTLKGHYNWVLCVSWS---PDGEVIATGSMDNTIRLWDPKSGQCLGDALRGHSK 230
Query 254 AVNQVAW--VNFCNPVC---LLSAGDDGVLLLW 281
+ ++W ++ P L S+ DG + +W
Sbjct 231 WITSLSWEPIHLVKPGSKPRLASSSKDGTIKIW 263
Score = 30.4 bits (67), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 27/109 (24%), Positives = 48/109 (44%), Gaps = 10/109 (9%)
Query 132 PRGNSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACP---SKICNQILISAATTPPV 188
P G ++ D IKL+D ++ F ++ + V + S C L+ + +
Sbjct 412 PDGRYIVSASFDNSIKLWD--GRDGKFISTFRGHVASVYQVAWSSDCR--LLVSCSKDTT 467
Query 189 LRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIW 237
L++WD+ RK+ GHK + + G R V SG +D + +W
Sbjct 468 LKVWDVRTRKLSVDLPGHKDEVYTVDWSVDGKR---VCSGGKDKMVRLW 513
> At5g08390
Length=823
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 54/106 (50%), Gaps = 7/106 (6%)
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWH 238
L++A ++LWDL+E K+V+ GH+ +++ P E SGS D + IW
Sbjct 166 LVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFH---PFGEFFASGSLDTNLKIWD 222
Query 239 -RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
R GC + +GH VN + + + ++S G+D V+ +W L
Sbjct 223 IRKKGC-IHTYKGHTRGVNVLRFT--PDGRWIVSGGEDNVVKVWDL 265
Score = 46.2 bits (108), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 26/98 (26%), Positives = 45/98 (45%), Gaps = 7/98 (7%)
Query 189 LRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQL 248
L++WD+ ++ + YKGH G +R P ++SG ED + +W G +L +
Sbjct 218 LKIWDIRKKGCIHTYKGHTRGVNVLRFT---PDGRWIVSGGEDNVVKVWDLTAGKLLHEF 274
Query 249 RGHAAAVNQVAWVNFCNPVCLLSAGD-DGVLLLWSLPT 285
+ H + ++F LL+ G D + W L T
Sbjct 275 KSHEGKIQS---LDFHPHEFLLATGSADKTVKFWDLET 309
Score = 31.2 bits (69), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 16/52 (30%), Positives = 27/52 (51%), Gaps = 7/52 (13%)
Query 188 VLRLWDLDERKIVQKYKGH--KVGRLAIRPAFGGPREELVISGSEDAQIYIW 237
V+++WDL K++ ++K H K+ L P E L+ +GS D + W
Sbjct 259 VVKVWDLTAGKLLHEFKSHEGKIQSLDFH-----PHEFLLATGSADKTVKFW 305
Score = 31.2 bits (69), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 17/67 (25%), Positives = 34/67 (50%), Gaps = 2/67 (2%)
Query 41 LDWDSTGKLLAAGGEAAFIEAWE--EGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRT 98
L + G+ + +GGE ++ W+ G+ + ++ H G I +L + P L + + D+T
Sbjct 242 LRFTPDGRWIVSGGEDNVVKVWDLTAGKLLHEFKSHEGKIQSLDFHPHEFLLATGSADKT 301
Query 99 MTLVSLE 105
+ LE
Sbjct 302 VKFWDLE 308
> 7300706
Length=510
Score = 52.0 bits (123), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 53/237 (22%), Positives = 101/237 (42%), Gaps = 23/237 (9%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ + +IS ++D T+ + + + T + E +L R G ++
Sbjct 249 HTGSVLCLQY--DDKVIISGSSDSTVRVWDVNTGEMVNTLIHHCEAVLHLRFNNGMMVTC 306
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWD 193
DR I ++D+ + E AA ++ I +A+ +++W
Sbjct 307 S-------KDRSIAVWDMTSPSEITLRRVLVGHRAAVNVVDFDEKYIVSASGDRTIKVWS 359
Query 194 LDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAA 253
+ V+ GHK G ++ R+ LV+SGS D I +W G L+ L GH
Sbjct 360 TSSCEFVRTLNGHKRGIACLQY-----RDRLVVSGSSDNSIRLWDIECGACLRVLEGHEE 414
Query 254 AVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSSPETATLSPPDQNT 310
V + + + ++S DG + +W L ++P ++ T L+ ++T
Sbjct 415 LVRCIRF----DTKRIVSGAYDGKIKVWDLVAA-----LDPRAASNTLCLNTLVEHT 462
> Hs14149684
Length=364
Score = 51.6 bits (122), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 49/259 (18%), Positives = 103/259 (39%), Gaps = 21/259 (8%)
Query 29 WRVQRLASATAFLDWDSTGKLLAAGGEAAFIEAWEEGRRIGD---YCPHGGSIVALQWVP 85
+R A +++ +G LLA+G + W + G+ + H ++ ++ +
Sbjct 12 YRFTGHKDAVTCVNFSPSGHLLASGSRDKTVRIWVPNVK-GESTVFRAHTATVRSVHFCS 70
Query 86 GSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADRQ 145
+ ++ + D+T+ + + + W+ + P G ++ D+
Sbjct 71 DGQSFVTASDDKTVKVWATHRQKFLFSLSQHINWVRCAK-----FSPDGRLIVSASDDKT 125
Query 146 IKLFDLVTKEETFWASEKDLVIAAC---PSKICNQILISAATTPPVLRLWDLDERKIVQK 202
+KL+D ++E E + PS C I+AA +++WD+ +++Q
Sbjct 126 VKLWDKSSRECVHSYCEHGGFVTYVDFHPSGTC----IAAAGMDNTVKVWDVRTHRLLQH 181
Query 203 YKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVN 262
Y+ H A+ P +I+ S D+ + I + G +L L GH VA+
Sbjct 182 YQLHSA---AVNGLSFHPSGNYLITASSDSTLKILDLMEGRLLYTLHGHQGPATTVAFSR 238
Query 263 FCNPVCLLSAGDDGVLLLW 281
S G D +++W
Sbjct 239 TGE--YFASGGSDEQVMVW 255
> Hs16117783
Length=605
Score = 51.2 bits (121), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 52/218 (23%), Positives = 93/218 (42%), Gaps = 34/218 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ R +I+ ++D T+ + + + T + E +L R G ++
Sbjct 342 HTGSVLCLQY--DERVIITGSSDSTVRVWDVNTGEMLNTLIHHCEAVLHLRFNNGMMVTC 399
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVI--------AACPSKICNQILISAATT 185
DR I ++D+ AS D+ + AA + I +A+
Sbjct 400 SK-------DRSIAVWDM--------ASPTDITLRRVLVGHRAAVNVVDFDDKYIVSASG 444
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
+++W+ + V+ GHK G ++ R+ LV+SGS D I +W G L
Sbjct 445 DRTIKVWNTSTCEFVRTLNGHKRGIACLQY-----RDRLVVSGSSDNTIRLWDIECGACL 499
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+ L GH V + + N ++S DG + +W L
Sbjct 500 RVLEGHEELVRCIRFDN----KRIVSGAYDGKIKVWDL 533
> Hs4502477
Length=569
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 52/218 (23%), Positives = 93/218 (42%), Gaps = 34/218 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ R +I+ ++D T+ + + + T + E +L R G ++
Sbjct 306 HTGSVLCLQY--DERVIITGSSDSTVRVWDVNTGEMLNTLIHHCEAVLHLRFNNGMMVTC 363
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVI--------AACPSKICNQILISAATT 185
DR I ++D+ AS D+ + AA + I +A+
Sbjct 364 SK-------DRSIAVWDM--------ASPTDITLRRVLVGHRAAVNVVDFDDKYIVSASG 408
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
+++W+ + V+ GHK G ++ R+ LV+SGS D I +W G L
Sbjct 409 DRTIKVWNTSTCEFVRTLNGHKRGIACLQY-----RDRLVVSGSSDNTIRLWDIECGACL 463
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+ L GH V + + N ++S DG + +W L
Sbjct 464 RVLEGHEELVRCIRFDN----KRIVSGAYDGKIKVWDL 497
> SPAC6B12.15
Length=314
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 39/151 (25%), Positives = 72/151 (47%), Gaps = 10/151 (6%)
Query 143 DRQIKLFDLVTKEETFWASEKDLV-----IAACPSKICNQILISAATTPPVLRLWDLDER 197
D+ I L++LV + + +++ L ++ C + +SA+ +RLWDL++
Sbjct 37 DKSIILWNLVRDDVNYGVAQRRLTGHSHFVSDCALSFDSHYALSASW-DKTIRLWDLEKG 95
Query 198 KIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLR-GHAAAVN 256
+ ++ GH L++ + P V+SGS D I IW+ + C GH+ V+
Sbjct 96 ECTHQFVGHTSDVLSVSIS---PDNRQVVSGSRDKTIKIWNIIGNCKYTITDGGHSDWVS 152
Query 257 QVAWVNFCNPVCLLSAGDDGVLLLWSLPTTS 287
V + + + +SAG D + +W L T S
Sbjct 153 CVRFSPNPDNLTFVSAGWDKAVKVWDLETFS 183
Score = 31.6 bits (70), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 27/101 (26%), Positives = 50/101 (49%), Gaps = 8/101 (7%)
Query 223 ELVISGSEDAQIYIWHRV-----WGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGV 277
++++SGS D I +W+ V +G ++L GH+ V+ A ++F + LSA D
Sbjct 29 DILLSGSRDKSIILWNLVRDDVNYGVAQRRLTGHSHFVSDCA-LSF-DSHYALSASWDKT 86
Query 278 LLLWSLPTTS-NEERVEPSSSPETATLSPPDQNTPDASSDQ 317
+ LW L + V +S + ++SP ++ S D+
Sbjct 87 IRLWDLEKGECTHQFVGHTSDVLSVSISPDNRQVVSGSRDK 127
> YLR129w
Length=943
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 31/107 (28%), Positives = 52/107 (48%), Gaps = 6/107 (5%)
Query 179 LISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWH 238
L++ V+++WDL + ++ + GHK ++ G R +ISGS+D+ I +W
Sbjct 93 LLAVGYADGVIKVWDLMSKTVLLNFNGHKAAITLLQFDGTGTR---LISGSKDSNIIVWD 149
Query 239 RVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPT 285
V L +LR H ++ W L+S DG++ LW L T
Sbjct 150 LVGEVGLYKLRSHKDSITGF-WCQ--GEDWLISTSKDGMIKLWDLKT 193
Score = 32.7 bits (73), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 24/81 (29%), Positives = 35/81 (43%), Gaps = 3/81 (3%)
Query 36 SATAFLDWDSTGKLLAAGGEAAFIEAWEEGRRIGDYC--PHGGSIVALQWVPGSRRLISM 93
+A L +D TG L +G + + I W+ +G Y H SI W G LIS
Sbjct 122 AAITLLQFDGTGTRLISGSKDSNIIVWDLVGEVGLYKLRSHKDSITGF-WCQGEDWLIST 180
Query 94 ATDRTMTLVSLEESSLTPTHV 114
+ D + L L+ TH+
Sbjct 181 SKDGMIKLWDLKTHQCIETHI 201
> 7302102
Length=647
Score = 50.1 bits (118), Expect = 6e-06, Method: Compositional matrix adjust.
Identities = 70/273 (25%), Positives = 114/273 (41%), Gaps = 33/273 (12%)
Query 32 QRLASATAFLDWDSTGKLLAAGGEAAFIEAWE-EGRRIGDYCPHGGSIVALQWVPGSRRL 90
++ AS L +D+ + +GG + + E +I ++ H G + L S L
Sbjct 144 EKHASNIFCLGFDTQNSYIFSGGNDDLVIQHDLETGKILNHFSHDGPVYGLSVDRISGHL 203
Query 91 ISMATDRTMTLV-SLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADRQIKLF 149
+S+AT+ LV L P + +++ P E F GN + A R L+
Sbjct 204 LSVATEHGEILVYDLRAGKSEPLAIAKFK--TPFNAVE-FHPLNGNFLATANAKRGAMLW 260
Query 150 DLVTKEETFWASEKDLVIAACPSKI-----CN--QILISAATTPPVLRLWDLDERKIVQK 202
DL ++ I PS + CN +L PP+L E +
Sbjct 261 DLRHHQQALCQYN---YIPESPSCMSVRFNCNGTLLLTLHRRLPPILYSPGAPE-PVATF 316
Query 203 YKGHKVGRLAIRP-AFGGPREELVISGSEDAQIYIWHRVWGCMLQQ-----------LRG 250
Y ++ F GP++ELV+SGS++ ++IW R+ G L + L G
Sbjct 317 YHDEYFNSCTMKSCTFAGPQDELVVSGSDNFNMFIW-RLEGVDLDEKNQWMETTPVILAG 375
Query 251 HAAAVNQVAWVNFCNPVCLL-SAGDDGVLLLWS 282
H + VNQ V + CLL S+G + ++ LWS
Sbjct 376 HRSIVNQ---VRYNRERCLLASSGVEKIIKLWS 405
> YMR116c
Length=319
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 65/129 (50%), Gaps = 9/129 (6%)
Query 182 AATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVW 241
+A+ LRLWD+ + Q++ GHK +++ + ++ISGS D I +W +
Sbjct 82 SASWDKTLRLWDVATGETYQRFVGHKSDVMSVDI---DKKASMIISGSRDKTIKVW-TIK 137
Query 242 GCMLQQLRGHAAAVNQVAWV----NFCNPVCLLSAGDDGVLLLWSLPTTSNE-ERVEPSS 296
G L L GH V+QV V + V ++SAG+D ++ W+L E + + +S
Sbjct 138 GQCLATLLGHNDWVSQVRVVPNEKADDDSVTIISAGNDKMVKAWNLNQFQIEADFIGHNS 197
Query 297 SPETATLSP 305
+ T T SP
Sbjct 198 NINTLTASP 206
> At5g16750
Length=876
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 30/118 (25%), Positives = 57/118 (48%), Gaps = 8/118 (6%)
Query 176 NQILISAATTPPVLRLWDLDERKIVQKYKGHK--VGRLAIRPAFGGPREELVISGSEDAQ 233
+++L SA + + R+WDL+ K ++ +KGH+ V +A + G L+ + D +
Sbjct 72 DKLLFSAGHSRQI-RVWDLETLKCIRSWKGHEGPVMGMACHASGG-----LLATAGADRK 125
Query 234 IYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEER 291
+ +W G RGH V+ + + N L+S DD + +W L + E++
Sbjct 126 VLVWDVDGGFCTHYFRGHKGVVSSILFHPDSNKNILISGSDDATVRVWDLNAKNTEKK 183
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 63/241 (26%), Positives = 101/241 (41%), Gaps = 27/241 (11%)
Query 84 VPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWI--LPGRPQEGFLLPRGNSVMIFF 141
V I+ A + +V +SS+ T E + + L P + L G+S
Sbjct 27 VSSDGSFIACACGDVINIVDSTDSSVKSTIEGESDTLTALALSPDDKLLFSAGHS----- 81
Query 142 ADRQIKLFDLVTKE--ETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKI 199
RQI+++DL T + ++ E ++ AC + +L +A VL +WD+D
Sbjct 82 --RQIRVWDLETLKCIRSWKGHEGPVMGMACHAS--GGLLATAGADRKVL-VWDVDGGFC 136
Query 200 VQKYKGHK--VGRLAIRPAFGGPREELVISGSEDAQIYIW----HRVWGCMLQQLRGHAA 253
++GHK V + P + ++ISGS+DA + +W L + H +
Sbjct 137 THYFRGHKGVVSSILFHP---DSNKNILISGSDDATVRVWDLNAKNTEKKCLAIMEKHFS 193
Query 254 AVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSSPETATLSPPDQNTPDA 313
AV +A + + L SAG D V+ LW L S + V E T TP A
Sbjct 194 AVTSIALSE--DGLTLFSAGRDKVVNLWDLHDYSCKATVATYEVLEAVT--TVSSGTPFA 249
Query 314 S 314
S
Sbjct 250 S 250
Score = 41.6 bits (96), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 42/186 (22%), Positives = 77/186 (41%), Gaps = 8/186 (4%)
Query 130 LLPRGNSVMIFFADRQIKLFDLVTK-EETFWASEKDLV-----IAACPSKICNQILISAA 183
+LP + ++ AD+Q + +V EET K LV IA + ++ A
Sbjct 318 MLPSDHGLLCVTADQQFFFYSVVENVEETELVLSKRLVGYNEEIADMKFLGDEEQFLAVA 377
Query 184 TTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGC 243
T +R++D+ GHK L++ L+++GS+D + +W+
Sbjct 378 TNLEEVRVYDVATMSCSYVLAGHKEVVLSLDTCVSSSGNVLIVTGSKDKTVRLWNATSKS 437
Query 244 MLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSSPETATL 303
+ GH + VA+ +S D L +WSL S E+ EP + + +
Sbjct 438 CIGVGTGHNGDILAVAFAKKSFSF-FVSGSGDRTLKVWSLDGIS-EDSEEPINLKTRSVV 495
Query 304 SPPDQN 309
+ D++
Sbjct 496 AAHDKD 501
Score = 41.2 bits (95), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 31/127 (24%), Positives = 58/127 (45%), Gaps = 8/127 (6%)
Query 160 ASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGG 219
A +KD+ A N L+ + +W L + V KGHK ++ +
Sbjct 497 AHDKDINSVAVAR---NDSLVCTGSEDRTASIWRLPDLVHVVTLKGHKRRIFSVEFS--- 550
Query 220 PREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLL 279
++ V++ S D + IW G L+ GH ++V + +++ + +S G DG+L
Sbjct 551 TVDQCVMTASGDKTVKIWAISDGSCLKTFEGHTSSVLRASFIT--DGTQFVSCGADGLLK 608
Query 280 LWSLPTT 286
LW++ T+
Sbjct 609 LWNVNTS 615
> 7293146
Length=340
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 50/216 (23%), Positives = 85/216 (39%), Gaps = 20/216 (9%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVV--EYEWILPGRPQEGFLL 131
H I A+ W SR L+S + D L+ + + H + W++
Sbjct 54 HLAKIYAMHWGNDSRNLVSASQDGK--LIVWDSHTTNKVHAIPLRSSWVMTCA-----YA 106
Query 132 PRGNSVMIFFADRQIKLFDLVTKEETFWASEK----DLVIAACPSKICNQILISAATTPP 187
P G+ V D +++L T+E S + ++ C NQI+ S+
Sbjct 107 PSGSYVACGGLDNMCSIYNLKTREGNVRVSRELPGHGGYLSCCRFLDDNQIVTSSGDMS- 165
Query 188 VLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQ 247
LWD++ V + GH +A+ A P+ + +SG+ DA +W G Q
Sbjct 166 -CGLWDIETGLQVTSFLGHTGDVMALSLA---PQCKTFVSGACDASAKLWDIREGVCKQT 221
Query 248 LRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
GH + +N V + F N + DD L+ +
Sbjct 222 FPGHESDINAVTF--FPNGQAFATGSDDATCRLFDI 255
Score = 30.0 bits (66), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 18/52 (34%), Positives = 27/52 (51%), Gaps = 2/52 (3%)
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSS 297
+ LRGH A + + W N + L+SA DG L++W TT+ + SS
Sbjct 49 RTLRGHLAKIYAMHWGN--DSRNLVSASQDGKLIVWDSHTTNKVHAIPLRSS 98
> Hs4504053
Length=340
Score = 49.7 bits (117), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 51/218 (23%), Positives = 85/218 (38%), Gaps = 24/218 (11%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVV--EYEWILPGRPQEGFLL 131
H I A+ W S+ L+S + D L+ + + H + W++
Sbjct 54 HLAKIYAMHWATDSKLLVSASQDGK--LIVWDSYTTNKVHAIPLRSSWVMTCA-----YA 106
Query 132 PRGNSVMIFFADRQIKLFDLVTKEETFWASEK----DLVIAACPSKICNQILISAATTPP 187
P GN V D +++L ++E S + ++ C N I+ S+ T
Sbjct 107 PSGNFVACGGLDNMCSIYNLKSREGNVKVSRELSAHTGYLSCCRFLDDNNIVTSSGDTTC 166
Query 188 VLRLWDLDERKIVQKYKGH--KVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
L WD++ + + GH LA+ P F L ISG+ DA +W G
Sbjct 167 AL--WDIETGQQKTVFVGHTGDCMSLAVSPDF-----NLFISGACDASAKLWDVREGTCR 219
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
Q GH + +N + + F N + + DD L+ L
Sbjct 220 QTFTGHESDINAICF--FPNGEAICTGSDDASCRLFDL 255
> Hs19913371
Length=514
Score = 49.7 bits (117), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 56/240 (23%), Positives = 87/240 (36%), Gaps = 40/240 (16%)
Query 41 LDWDSTGKLLAAGGEAAFIEAW-EEGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTM 99
LDW+S G LLA G F W ++G H G I AL+W ++S D+T
Sbjct 231 LDWNSEGTLLATGSYDGFARIWTKDGNLASTLGQHKGPIFALKWNKKGNFILSAGVDKTT 290
Query 100 TL---VSLEESSLTPTH---VVEYEWILPGRPQEGFLLPRGNSVMIFFA-----DRQIKL 148
+ + E P H ++ +W Q ++ M DR IK
Sbjct 291 IIWDAHTGEAKQQFPFHSAPALDVDW------QSNNTFASCSTDMCIHVCKLGQDRPIKT 344
Query 149 FDLVTKEETF--WASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGH 206
F T E W +L +A+C + L++W + + V + H
Sbjct 345 FQGHTNEVNAIKWDPTGNL-LASCSDDM-------------TLKIWSMKQDNCVHDLQAH 390
Query 207 KVGRLAIR-----PAFGGPREELVI-SGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAW 260
I+ P P L++ S S D+ + +W G + L H V VA+
Sbjct 391 NKEIYTIKWSPTGPGTNNPNANLMLASASFDSTVRLWDVDRGICIHTLTKHQEPVYSVAF 450
> Hs15150805
Length=522
Score = 49.7 bits (117), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 55/238 (23%), Positives = 86/238 (36%), Gaps = 36/238 (15%)
Query 41 LDWDSTGKLLAAGGEAAFIEAW-EEGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTM 99
LDW+S G LLA G F W E G H G I AL+W ++S D+T
Sbjct 241 LDWNSDGTLLAMGSYDGFARIWTENGNLASTLGQHKGPIFALKWNKKGNYVLSAGVDKTT 300
Query 100 TL---VSLEESSLTPTH---VVEYEWILPGRPQEGFLLPRGNSVMIFFA-----DRQIKL 148
+ + E P H ++ +W Q ++ M D +K
Sbjct 301 IIWDAHTGEAKQQFPFHSAPALDVDW------QNNMTFASCSTDMCIHVCRLGCDHPVKT 354
Query 149 FDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKV 208
F T E I PS +L+++ + L++W + + V + H
Sbjct 355 FQGHTNEVN--------AIKWDPSG----MLLASCSDDMTLKIWSMKQDACVHDLQAHSK 402
Query 209 GRLAIR-----PAFGGPREELVI-SGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAW 260
I+ PA P +++ S S D+ + +W G L H V VA+
Sbjct 403 EIYTIKWSPTGPATSNPNSSIMLASASFDSTVRLWDVEQGVCTHTLMKHQEPVYSVAF 460
> Hs5032159
Length=577
Score = 49.7 bits (117), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 54/238 (22%), Positives = 88/238 (36%), Gaps = 36/238 (15%)
Query 41 LDWDSTGKLLAAGGEAAFIEAW-EEGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTM 99
LDW++ G LLA G F W E+G H G I AL+W ++S D+T
Sbjct 294 LDWNTNGTLLATGSYDGFARIWTEDGNLASTLGQHKGPIFALKWNRKGNYILSAGVDKTT 353
Query 100 TL---VSLEESSLTPTH---VVEYEWILPGRPQEGFLLPRGNSVMIFF-----ADRQIKL 148
+ + E P H ++ +W Q ++ M DR +K
Sbjct 354 IIWDAHTGEAKQQFPFHSAPALDVDW------QNNTTFASCSTDMCIHVCRLGCDRPVKT 407
Query 149 FDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKV 208
F T E I PS +L+++ + L++W + + + + H
Sbjct 408 FQGHTNEVN--------AIKWDPSG----MLLASCSDDMTLKIWSMKQEVCIHDLQAHNK 455
Query 209 GRLAIR-----PAFGGPREELVI-SGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAW 260
I+ PA P +++ S S D+ + +W G L H V VA+
Sbjct 456 EIYTIKWSPTGPATSNPNSNIMLASASFDSTVRLWDIERGVCTHTLTKHQEPVYSVAF 513
> Hs4505895
Length=514
Score = 49.7 bits (117), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 46/157 (29%), Positives = 66/157 (42%), Gaps = 10/157 (6%)
Query 143 DRQIKLFDLVTKEETFWASEKDLVIAACPSKICN-QILISAATTPPVLRLWDLDERKIVQ 201
D +++D+ TK S +A + QI+ + T +RLWDL K
Sbjct 309 DSTARIWDVRTKASVHTLSGHTNAVATVRCQAAEPQIITGSHDT--TIRLWDLVAGKTRV 366
Query 202 KYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWV 261
HK ++R PR SGS D I W G +Q L GH A +N + V
Sbjct 367 TLTNHK---KSVRAVVLHPRHYTFASGSPD-NIKQWKFPDGSFIQNLSGHNAIINTLT-V 421
Query 262 NFCNPVCLLSAGDDGVLLLWSLPTTSNEERVEPSSSP 298
N + L+S D+G + LW T N +RV + P
Sbjct 422 N--SDGVLVSGADNGTMHLWDWRTGYNFQRVHAAVQP 456
Score = 38.5 bits (88), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 35/146 (23%), Positives = 59/146 (40%), Gaps = 14/146 (9%)
Query 142 ADRQIKLFDLVTKEETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDERKIVQ 201
ADR IK++DL + + + + + L S V + WDL+ K+++
Sbjct 224 ADRTIKIWDLASGKLKLSLTGHISTVRGVIVSTRSPYLFSCGEDKQV-KCWDLEYNKVIR 282
Query 202 KYKGH--KVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVA 259
Y GH V L + P +++++ S D+ IW + L GH AV V
Sbjct 283 HYHGHLSAVYGLDLHPTI-----DVLVTCSRDSTARIWDVRTKASVHTLSGHTNAVATVR 337
Query 260 WVNFCNPV--CLLSAGDDGVLLLWSL 283
C +++ D + LW L
Sbjct 338 ----CQAAEPQIITGSHDTTIRLWDL 359
Score = 30.0 bits (66), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 2/64 (3%)
Query 220 PREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLL 279
P + ++GS D I IW G + L GH + V V V+ +P L S G+D +
Sbjct 214 PGNQWFVTGSADRTIKIWDLASGKLKLSLTGHISTVRGVI-VSTRSPY-LFSCGEDKQVK 271
Query 280 LWSL 283
W L
Sbjct 272 CWDL 275
> SPAC18B11.10
Length=614
Score = 49.3 bits (116), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 44/158 (27%), Positives = 74/158 (46%), Gaps = 14/158 (8%)
Query 132 PRGNSVMIFFADRQIKLFDLVTKE--ETFWASEKDLVIAACPSKICNQILISAATTPPVL 189
P G ++ DRQIKL+DL T++ F E+D+ N I + +
Sbjct 370 PDGKYLVTGTEDRQIKLWDLSTQKVRYVFSGHEQDIYSLDFSH---NGRFIVSGSGDRTA 426
Query 190 RLWDLDERKIVQKYK-GHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQL 248
RLWD++ + + K + + V +AI P ++ + GS D I +W V G ++++L
Sbjct 427 RLWDVETGQCILKLEIENGVTAIAI-----SPNDQFIAVGSLDQIIRVW-SVSGTLVERL 480
Query 249 RGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSLPTT 286
GH +V +A+ + LLS D + +W L T
Sbjct 481 EGHKESVYSIAFS--PDSSILLSGSLDKTIKVWELQAT 516
> Hs16306496
Length=529
Score = 49.3 bits (116), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 51/218 (23%), Positives = 92/218 (42%), Gaps = 34/218 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ R +++ ++D T+ + + + T + E +L R G ++
Sbjct 266 HTGSVLCLQY--DERVIVTGSSDSTVRVWDVNTGEVLNTLIHHNEAVLHLRFSNGLMVTC 323
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVI--------AACPSKICNQILISAATT 185
DR I ++D+ AS D+ + AA + I +A+
Sbjct 324 SK-------DRSIAVWDM--------ASATDITLRRVLVGHRAAVNVVDFDDKYIVSASG 368
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
+++W + V+ GHK G ++ R+ LV+SGS D I +W G L
Sbjct 369 DRTIKVWSTSTCEFVRTLNGHKRGIACLQY-----RDRLVVSGSSDNTIRLWDIECGACL 423
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+ L GH V + + N ++S DG + +W L
Sbjct 424 RVLEGHEELVRCIRFDN----KRIVSGAYDGKIKVWDL 457
> Hs16306498
Length=508
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 51/218 (23%), Positives = 92/218 (42%), Gaps = 34/218 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ R +++ ++D T+ + + + T + E +L R G ++
Sbjct 245 HTGSVLCLQY--DERVIVTGSSDSTVRVWDVNTGEVLNTLIHHNEAVLHLRFSNGLMVTC 302
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVI--------AACPSKICNQILISAATT 185
DR I ++D+ AS D+ + AA + I +A+
Sbjct 303 SK-------DRSIAVWDM--------ASATDITLRRVLVGHRAAVNVVDFDDKYIVSASG 347
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
+++W + V+ GHK G ++ R+ LV+SGS D I +W G L
Sbjct 348 DRTIKVWSTSTCEFVRTLNGHKRGIACLQY-----RDRLVVSGSSDNTIRLWDIECGACL 402
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+ L GH V + + N ++S DG + +W L
Sbjct 403 RVLEGHEELVRCIRFDN----KRIVSGAYDGKIKVWDL 436
> Hs16306494
Length=542
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 51/218 (23%), Positives = 92/218 (42%), Gaps = 34/218 (15%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPR 133
H GS++ LQ+ R +++ ++D T+ + + + T + E +L R G ++
Sbjct 279 HTGSVLCLQY--DERVIVTGSSDSTVRVWDVNTGEVLNTLIHHNEAVLHLRFSNGLMVTC 336
Query 134 GNSVMIFFADRQIKLFDLVTKEETFWASEKDLVI--------AACPSKICNQILISAATT 185
DR I ++D+ AS D+ + AA + I +A+
Sbjct 337 SK-------DRSIAVWDM--------ASATDITLRRVLVGHRAAVNVVDFDDKYIVSASG 381
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCML 245
+++W + V+ GHK G ++ R+ LV+SGS D I +W G L
Sbjct 382 DRTIKVWSTSTCEFVRTLNGHKRGIACLQY-----RDRLVVSGSSDNTIRLWDIECGACL 436
Query 246 QQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+ L GH V + + N ++S DG + +W L
Sbjct 437 RVLEGHEELVRCIRFDN----KRIVSGAYDGKIKVWDL 470
> Hs11055998
Length=340
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 49/216 (22%), Positives = 85/216 (39%), Gaps = 20/216 (9%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVV--EYEWILPGRPQEGFLL 131
H I A+ W SR L+S + D L+ + + H + W++
Sbjct 54 HLAKIYAMHWGYDSRLLVSASQDGK--LIIWDSYTTNKMHAIPLRSSWVMTCA-----YA 106
Query 132 PRGNSVMIFFADRQIKLFDLVTKEETFWASEK----DLVIAACPSKICNQILISAATTPP 187
P GN V D +++L T+E S + ++ C +QI+ S+ T
Sbjct 107 PSGNYVACGGLDNICSIYNLKTREGNVRVSRELPGHTGYLSCCRFLDDSQIVTSSGDTTC 166
Query 188 VLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQ 247
L WD++ + + GH +++ + P +SG+ DA +W G Q
Sbjct 167 AL--WDIETAQQTTTFTGHSGDVMSLSLS---PDMRTFVSGACDASSKLWDIRDGMCRQS 221
Query 248 LRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
GH + +N V++ F N + DD L+ L
Sbjct 222 FTGHVSDINAVSF--FPNGYAFATGSDDATCRLFDL 255
> YCR084c
Length=713
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 62/239 (25%), Positives = 99/239 (41%), Gaps = 34/239 (14%)
Query 47 GKLLAAGGEAAFIEAWE-EGRRIGDYCP-HGGSIVALQWVPGSRRLISMATDRTMTLVSL 104
GK LA G E I W+ E R+I H I +L + P +L+S + DRT+ + L
Sbjct 455 GKFLATGAEDRLIRIWDIENRKIVMILQGHEQDIYSLDYFPSGDKLVSGSGDRTVRIWDL 514
Query 105 EESSLTPTHVVEYE-WILPGRPQEGFLLPRGNSVMIFFADRQIKLFDLVT--------KE 155
+ T +E + P +G + G+ DR ++++D T E
Sbjct 515 RTGQCSLTLSIEDGVTTVAVSPGDGKYIAAGS------LDRAVRVWDSETGFLVERLDSE 568
Query 156 ETFWASEKDLVIAACPSKICNQILISAATTPPVLRLWDLDE--RKIVQK----------Y 203
KD V + ++ Q ++S + V +LW+L K K Y
Sbjct 569 NESGTGHKDSVYSVVFTR-DGQSVVSGSLDRSV-KLWNLQNANNKSDSKTPNSGTCEVTY 626
Query 204 KGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVN 262
GHK L++ +E ++SGS+D + W + G L L+GH +V VA N
Sbjct 627 IGHKDFVLSVATT---QNDEYILSGSKDRGVLFWDKKSGNPLLMLQGHRNSVISVAVAN 682
> Hs11321585
Length=340
Score = 49.3 bits (116), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 51/216 (23%), Positives = 85/216 (39%), Gaps = 20/216 (9%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVV--EYEWILPGRPQEGFLL 131
H I A+ W SR L+S + D L+ + + H + W++
Sbjct 54 HLAKIYAMHWGTDSRLLVSASQDGK--LIIWDSYTTNKVHAIPLRSSWVMTCA-----YA 106
Query 132 PRGNSVMIFFADRQIKLFDLVTKEETFWASEK----DLVIAACPSKICNQILISAATTPP 187
P GN V D +++L T+E S + ++ C NQI+ S+ T
Sbjct 107 PSGNYVACGGLDNICSIYNLKTREGNVRVSRELAGHTGYLSCCRFLDDNQIVTSSGDTTC 166
Query 188 VLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYIWHRVWGCMLQQ 247
L WD++ + + GH +++ A P L +SG+ DA +W G Q
Sbjct 167 AL--WDIETGQQTTTFTGHTGDVMSLSLA---PDTRLFVSGACDASAKLWDVREGMCRQT 221
Query 248 LRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
GH + +N + + F N + DD L+ L
Sbjct 222 FTGHESDINAICF--FPNGNAFATGSDDATCRLFDL 255
Score = 32.3 bits (72), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/96 (23%), Positives = 37/96 (38%), Gaps = 11/96 (11%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPT---HVVEYEWILPGRPQEGFL 130
H G +++L P +R +S A D + L + E T H + I
Sbjct 183 HTGDVMSLSLAPDTRLFVSGACDASAKLWDVREGMCRQTFTGHESDINAIC--------F 234
Query 131 LPRGNSVMIFFADRQIKLFDLVTKEETFWASEKDLV 166
P GN+ D +LFDL +E S +++
Sbjct 235 FPNGNAFATGSDDATCRLFDLRADQELMTYSHDNII 270
> 7296209
Length=700
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 55/255 (21%), Positives = 94/255 (36%), Gaps = 22/255 (8%)
Query 41 LDWDSTGKLLAAGGEAAFIEAWE-EGRRIGDYCPHGGSIVALQWVPGSRRLISMATDRTM 99
LDW+ G LLA G + W+ +GR H G I AL+W ++S D+T
Sbjct 416 LDWNCDGSLLATGSYDGYARIWKTDGRLASTLGQHKGPIFALKWNKCGNYILSAGVDKTT 475
Query 100 TLVSLEESSLTPTHVVEYEWILPGRPQEGFLLPRGNSVMIFFADRQIKLFDLVTKE--ET 157
+ T ++ P + D++I + L E +T
Sbjct 476 IIWDASTGQCT------QQFAFHSAPALDVDWQTNQAFASCSTDQRIHVCRLGVNEPIKT 529
Query 158 FWASEKDL-VIAACPSKICNQILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIR-- 214
F ++ I CP L+++ + L++W ++ + + H I+
Sbjct 530 FKGHTNEVNAIKWCPQG----QLLASCSDDMTLKIWSMNRDRCCHDLQAHSKEIYTIKWS 585
Query 215 ---PAFGGPREELVI-SGSEDAQIYIWHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLL 270
P P L++ S S D+ + +W G + L H V VA+ + L
Sbjct 586 PTGPGTNNPNTNLILASASFDSTVRLWDVERGSCIHTLTKHTEPVYSVAFS--PDGKHLA 643
Query 271 SAGDDGVLLLWSLPT 285
S D + +WS T
Sbjct 644 SGSFDKCVHIWSTQT 658
Score = 33.1 bits (74), Expect = 0.81, Method: Compositional matrix adjust.
Identities = 25/104 (24%), Positives = 47/104 (45%), Gaps = 5/104 (4%)
Query 177 QILISAATTPPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPREELVISGSEDAQIYI 236
+++++A+ +RLWD++ + H ++ AF P + + SGS D ++I
Sbjct 597 NLILASASFDSTVRLWDVERGSCIHTLTKHTEPVYSV--AFS-PDGKHLASGSFDKCVHI 653
Query 237 WHRVWGCMLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLL 280
W G ++ +G + +V W N SA D V +L
Sbjct 654 WSTQTGQLVHSYKG-TGGIFEVCW-NSKGTKVGASASDGSVFVL 695
> Hs5729852
Length=353
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 47/220 (21%), Positives = 88/220 (40%), Gaps = 23/220 (10%)
Query 74 HGGSIVALQWVPGSRRLISMATDRTMTLVSLEESSLTPTHVVEY--EWILPGRPQEGFLL 131
HG ++ + W RR++S + D ++ + + H V W++
Sbjct 62 HGNKVLCMDWCKDKRRIVSSSQDG--KVIVWDSFTTNKEHAVTMPCTWVMACA-----YA 114
Query 132 PRGNSVMIFFADRQIKLFDLV-TKEETFWASEKDLV-----IAACPSKICNQILISAATT 185
P G ++ D + ++ L K E A +K + ++AC S + + I A+
Sbjct 115 PSGCAIACGGLDNKCSVYPLTFDKNENMAAKKKSVAMHTNYLSAC-SFTNSDMQILTASG 173
Query 186 PPVLRLWDLDERKIVQKYKGHKVGRLAIRPAFGGPRE--ELVISGSEDAQIYIWHRVWGC 243
LWD++ +++Q + GH L + A P E +SG D + +W G
Sbjct 174 DGTCALWDVESGQLLQSFHGHGADVLCLDLA---PSETGNTFVSGGCDKKAMVWDMRSGQ 230
Query 244 MLQQLRGHAAAVNQVAWVNFCNPVCLLSAGDDGVLLLWSL 283
+Q H + +N V + + + S DD L+ L
Sbjct 231 CVQAFETHESDINSVRY--YPSGDAFASGSDDATCRLYDL 268
Lambda K H
0.319 0.135 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7378771334
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40