bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
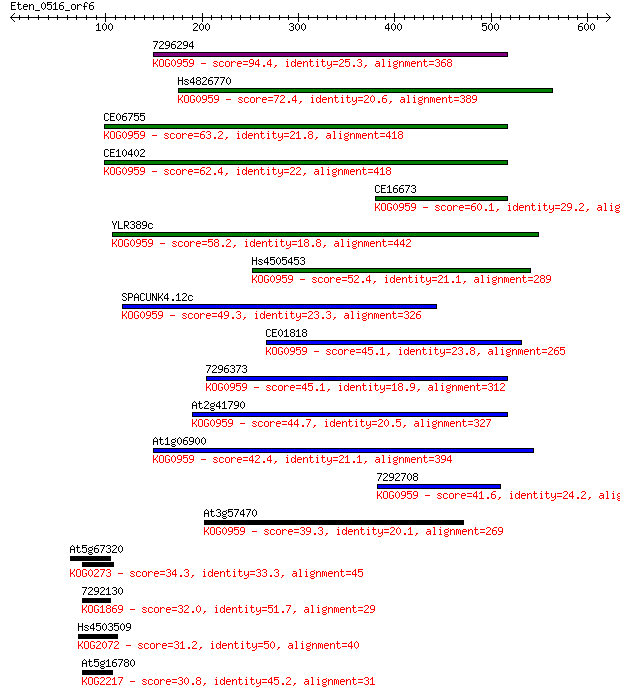
Query= Eten_0516_orf6
Length=623
Score E
Sequences producing significant alignments: (Bits) Value
7296294 94.4 6e-19
Hs4826770 72.4 3e-12
CE06755 63.2 2e-09
CE10402 62.4 3e-09
CE16673 60.1 2e-08
YLR389c 58.2 6e-08
Hs4505453 52.4 3e-06
SPACUNK4.12c 49.3 2e-05
CE01818 45.1 5e-04
7296373 45.1 5e-04
At2g41790 44.7 7e-04
At1g06900 42.4 0.003
7292708 41.6 0.006
At3g57470 39.3 0.024
At5g67320 34.3 0.94
7292130 32.0 4.1
Hs4503509 31.2 7.9
At5g16780 30.8 8.7
> 7296294
Length=990
Score = 94.4 bits (233), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 93/385 (24%), Positives = 166/385 (43%), Gaps = 31/385 (8%)
Query 149 VWWQGQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITH 208
VW + P+ + S D L+ + + + + L E D + +
Sbjct 532 VWHKQDNQFNKPKACMTFDMSNPIAYLDPLNCNLNHMMVMLLKDQLNEYLYDAELASLKL 591
Query 209 SLAFKGTGFHMTFEGYTKTQ---LDKLMSHVASVLRDPSVVESERFERVKQKQIKLLADP 265
S+ K G T G++ Q L+KL+ H L D S+ E +RF+ +K++ ++ L +
Sbjct 592 SVMGKSCGIDFTIRGFSDKQVVLLEKLLDH----LFDFSIDE-KRFDILKEEYVRSLKNF 646
Query 266 ATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE-LKNVHVDAFVMGNI 324
++H++ A+L +A++ M+LL+A+E TYD + E + +H + F+ GN+
Sbjct 647 KAEQPYQHSIYYLALLLTENAWANMELLDAMELVTYDRVLNFAKEFFQRLHTECFIFGNV 706
Query 325 DRDQSLLLTESFLEQAG--FTPIEHDDAVESLAMEQKQTIEATLANPIKGD--------- 373
+ Q+ + AG T +E +A L + +Q ++ + GD
Sbjct 707 TKQQA-------TDIAGRVNTRLEATNA-SKLPILARQMLKKREYKLLAGDSYLFEKENE 758
Query 374 --KDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTA 431
K + + Q G + + + ++SQ L+ YD LRT+ QLGYI + +
Sbjct 759 FHKSSCAQLYLQCGAQTDHTNIMVNLVSQVLSEPCYDCLRTKEQLGYIVFSGVRKVNGAN 818
Query 432 LLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSED 491
++ V+ +K HP V I+ L I +MP E R KEA K + +
Sbjct 819 GIRIIVQSAK-HPSYVEDRIENFLQTYLQVIEDMPLDEFERHKEALAVKKLEKPKTIFQQ 877
Query 492 FKKSADEIFSHANCFSKRDLEVKYL 516
F + EI F + + EV L
Sbjct 878 FSQFYGEIAMQTYHFEREEAEVAIL 902
> Hs4826770
Length=1019
Score = 72.4 bits (176), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 80/399 (20%), Positives = 169/399 (42%), Gaps = 17/399 (4%)
Query 175 ADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYTKTQ---LDK 231
D L + L + + L E + G+++ L G +++ +GY Q L K
Sbjct 585 VDPLHCNMAYLYLELLKDSLNEYAYAAELAGLSYDLQNTIYGMYLSVKGYNDKQPILLKK 644
Query 232 LMSHVASVLRDPSVVESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMD 291
++ +A+ ++ +RFE +K+ ++ L + +HA+ +L A+++ +
Sbjct 645 IIEKMATF-----EIDEKRFEIIKEAYMRSLNNFRAEQPHQHAMYYLRLLMTEVAWTKDE 699
Query 292 LLNALEQTTYDDSIAKLSEL-KNVHVDAFVMGNIDRDQSL----LLTESFLEQAGFTPIE 346
L AL+ T A + +L +H++A + GNI + +L ++ ++ +E A P+
Sbjct 700 LKEALDDVTLPRLKAFIPQLLSRLHIEALLHGNITKQAALGIMQMVEDTLIEHAHTKPLL 759
Query 347 HDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRI 406
V ++ + + + +Q + S E + L + Q ++
Sbjct 760 PSQLVRYREVQLPDRGWFVYQQRNEVHNNCGIEIYYQTDMQSTSENMFLELFCQIISEPC 819
Query 407 YDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMP 466
+++LRT+ QLGYI + +A L+ F+ S+ P + ++A L + I +M
Sbjct 820 FNTLRTKEQLGYIVFSGPRRANGIQSLR-FIIQSEKPPHYLESRVEAFLITMEKSIEDMT 878
Query 467 EAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLL 526
E + +A + S + K EI S F + + EV YL +++ ++
Sbjct 879 EEAFQKHIQALAIRRLDKPKKLSAECAKYWGEIISQQYNFDRDNTEVAYLKT-LTKEDII 937
Query 527 RTFTKL--SDPSRRMVVKLIADLEPAKEVTLIGEAKGQD 563
+ + ++ D RR V + ++GE Q+
Sbjct 938 KFYKEMLAVDAPRRHKVSVHVLAREMDSCPVVGEFPCQN 976
> CE06755
Length=980
Score = 63.2 bits (152), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 91/446 (20%), Positives = 174/446 (39%), Gaps = 34/446 (7%)
Query 99 ERERERPNAFRMPPPLLHIPKASELEIL-----PGLLGLNE-PELISEQGGNAGTAVWWQ 152
E+ ++ NA + LH+P+ +E P NE P+LIS+ G + VW++
Sbjct 445 EKMKKYENALKTSHHALHLPEKNEYIATNFGQKPRESVKNEHPKLISDDGW---SRVWFK 501
Query 153 GQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAF 212
+P+ + + + + S+ L + L EET + G+
Sbjct 502 QDDEYNMPKQETKFALTTPIVSQNPRISLISSLWLWCFCDILSEETYNAALAGLGCQFEL 561
Query 213 KGTGFH----------------MTFEGYTKTQLDKLMSHVASVLRDPSVVESERFERVKQ 256
G + GY + Q + H+ S + + + + RFE + +
Sbjct 562 SPFGVQKQSTDGREAERHASLTLHVYGYDEKQ-PLFVKHLTSCMINFKI-DRTRFEVLFE 619
Query 257 KQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE-LKNVH 315
+ L + A + + +L + +S+ LL + T ++ E L+ H
Sbjct 620 SLKRTLTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLENVQGFAREMLQAFH 679
Query 316 VDAFVMGNIDRDQSLLLTESFLE-----QAGFTPIEHDDAVESLAMEQKQTIEATLANPI 370
++ FV GN +++ L++ ++ P+ ++ + E +
Sbjct 680 MELFVHGNSTEKEAIQLSKELMDILKSAAPNSRPLYRNEHNPRREFQLNNGDEYIYRHLQ 739
Query 371 KGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAAST 430
K V +Q+GV + + + ++ Q + ++D+LRT LGYI
Sbjct 740 KTHDAGCVEVTYQIGVQNKYDNAVVGLIDQLIKEPVFDTLRTNEALGYIVWTGCRFNCGA 799
Query 431 ALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSE 490
L FV+G KS D V++ I+ L + I MP+ E + A+L + S
Sbjct 800 VALNIFVQGPKS-VDYVLERIEVFLESVRKEIIEMPQDEFEKKVAGMIARLEEKPKTLSN 858
Query 491 DFKKSADEIFSHANCFSKRDLEVKYL 516
FK+ +I F++R+ EVK L
Sbjct 859 RFKRFWYQIECRQYDFARREKEVKVL 884
> CE10402
Length=1067
Score = 62.4 bits (150), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 92/445 (20%), Positives = 178/445 (40%), Gaps = 32/445 (7%)
Query 99 ERERERPNAFRMPPPLLHIPK-----ASELEILPGLLGLNE-PELISEQGGNAGTAVWWQ 152
E ++ NA + LH+P+ A+ + P NE P LIS+ G + VW++
Sbjct 531 ETMKKYENALKTSHHALHLPEKNEYIATNFDQKPRESVKNEHPRLISDDGW---SRVWFK 587
Query 153 GQGFSALPRVAVQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAF 212
+P+ +L+ + + S+ L +++ L EET + G+ L
Sbjct 588 QDDEYNMPKQETKLALTTPMVAQNPRMSLLSSLWLWCLSDTLAEETYNADLAGLKCQLES 647
Query 213 KGTGFHM--------------TFEGYTKTQLDKLMS-HVASVLRDPSVVESERFERVKQK 257
G M T Y + L + H+A+ + + + + RF+ + +
Sbjct 648 SPFGVQMRVSNRREAERHASLTLHVYGYDEKQALFAKHLANRMTNFKI-DKTRFDVLFES 706
Query 258 QIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE-LKNVHV 316
+ L + A + + +L + +S+ LL + T +D E L+ H+
Sbjct 707 LKRALTNHAFSQPYLLTQHYNQLLIVDKVWSKEQLLAVCDSVTLEDVQGFAKEMLQAFHM 766
Query 317 DAFVMGNIDRDQSLLLTESFLE-----QAGFTPIEHDDAVESLAMEQKQTIEATLANPIK 371
+ FV GN +++ L++ ++ P+ ++ ++ E + K
Sbjct 767 ELFVHGNSTEKEAIQLSKELMDVLKSAAPNSRPLYRNEHNPRRELQLNNGDEYVYRHLQK 826
Query 372 GDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTA 431
V +Q+GV + + + ++ Q + +++LRT LGYI T
Sbjct 827 THDVGCVEVTYQIGVQNTYDNAVVGLIDQLIREPAFNTLRTNEALGYIVWTGSRLNCGTV 886
Query 432 LLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSED 491
L V+G KS D V++ I+ L + I+ MP+ E A+L + S
Sbjct 887 ALNVIVQGPKS-VDHVLERIEVFLESVRKEIAEMPQEEFDNQVSGMIARLEEKPKTLSSR 945
Query 492 FKKSADEIFSHANCFSKRDLEVKYL 516
F++ +EI F++R+ EV L
Sbjct 946 FRRFWNEIECRQYNFARREEEVALL 970
> CE16673
Length=301
Score = 60.1 bits (144), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 40/137 (29%), Positives = 65/137 (47%), Gaps = 1/137 (0%)
Query 380 VQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG 439
V +Q+GV + + + ++ + +D+LRT+ LGYI + T LQ V+G
Sbjct 76 VTYQIGVQNTYDNAVIGLIKNLITEPAFDTLRTKESLGYIVWTRTHFNCGTVALQILVQG 135
Query 440 SKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEI 499
KS D V++ I+A L + I MP+ E A+L + S FKK DEI
Sbjct 136 PKS-VDHVLERIEAFLESVRKEIVEMPQEEFENRVSGLIAQLEEKPKTLSCRFKKFWDEI 194
Query 500 FSHANCFSKRDLEVKYL 516
F++ + +V+ L
Sbjct 195 ECRQYNFTRIEEDVELL 211
> YLR389c
Length=988
Score = 58.2 bits (139), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 83/452 (18%), Positives = 193/452 (42%), Gaps = 21/452 (4%)
Query 107 AFRMPPPLLHIPKASELEILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPRVAVQL 166
A +P P + +++ + G+ L+EP L+ + + +W++ PR + L
Sbjct 530 ALTLPRPNEFVSTNFKVDKIDGIKPLDEPVLLL---SDDVSKLWYKKDDRFWQPRGYIYL 586
Query 167 SGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYTK 226
S + A +++ ++ + L++ D + S G +T G+
Sbjct 587 SFKLPHTHASIINSMLSTLYTQLANDALKDVQYDAACADLRISFNKTNQGLAITASGFN- 645
Query 227 TQLDKLMSHVASVLRDPSVVE--SERFERVKQKQIKLLADPATNMAFEHALQAAAILTRN 284
+KL+ + L+ + E +RFE +K K I+ L + + + +
Sbjct 646 ---EKLIILLTRFLQGVNSFEPKKDRFEILKDKTIRHLKNLLYEVPYSQMSNYYNAIINE 702
Query 285 DAFSRMDLLNALEQTTYDDSIAKLSEL-KNVHVDAFVMGNIDRDQSL---LLTESFLEQA 340
++S + L E+ T++ I + + + V+ + + GNI +++L L +S +
Sbjct 703 RSWSTAEKLQVFEKLTFEQLINFIPTIYEGVYFETLIHGNIKHEEALEVDSLIKSLIPNN 762
Query 341 GFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHASLVQF--QLGVPSIEERVNLAIL 398
++ + S + + +T A +K ++ S +Q QL V S + +
Sbjct 763 IHNLQVSNNRLRSYLLPKGKTFRYETA--LKDSQNVNSCIQHVTQLDVYSEDLSALSGLF 820
Query 399 SQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKA 458
+Q ++ +D+LRT+ QLGY+ + TA ++ ++ + P + I+
Sbjct 821 AQLIHEPCFDTLRTKEQLGYVVFSSSLNNHGTANIRILIQSEHTTP-YLEWRINNFYETF 879
Query 459 KDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDN 518
+ +MPE + + KEA L + N +E+ + I+ F+ R + K + N
Sbjct 880 GQVLRDMPEEDFEKHKEALCNSLLQKFKNMAEESARYTAAIYLGDYNFTHRQKKAKLVAN 939
Query 519 DFSRKHLLRTFTK--LSDPSRRMVVKLIADLE 548
+++ ++ + +S+ + ++++ L + +E
Sbjct 940 -ITKQQMIDFYENYIMSENASKLILHLKSQVE 970
> Hs4505453
Length=1219
Score = 52.4 bits (124), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 61/295 (20%), Positives = 128/295 (43%), Gaps = 13/295 (4%)
Query 252 ERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNAL-EQTTYDDSIAKLSE 310
E++K+ +L P T A ++ +S +D AL + + + ++ + E
Sbjct 864 EQLKKTYFNILIKPET-----LAKDVRLLILEYARWSMIDKYQALMDGLSLESLLSFVKE 918
Query 311 LKN-VHVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANP 369
K+ + V+ V GN+ +S+ + +++ F P+E + V+ +E
Sbjct 919 FKSQLFVEGLVQGNVTSTESMDFLKYVVDKLNFKPLEQEMPVQFQVVELPSGHHLCKVKA 978
Query 370 I-KGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGY--IAGAKESQ 426
+ KGD + V +Q G S+ E + +L + +D LRT+ LGY + +
Sbjct 979 LNKGDANSEVTVYYQSGTRSLREYTLMELLVMHMEEPCFDFLRTKQTLGYHVYPTCRNTS 1038
Query 427 AASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVE- 485
+ + +K + + V K I+ L+ ++ I N+ E + + A KL + E
Sbjct 1039 GILGFSVTVGTQATKYNSEVVDKKIEEFLSSFEEKIENLTEEAFNT-QVTALIKLKECED 1097
Query 486 ANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDPSRRMV 540
+ E+ ++ +E+ + F + E++ L + FS+ L+ F P +M+
Sbjct 1098 THLGEEVDRNWNEVVTQQYLFDRLAHEIEALKS-FSKSDLVNWFKAHRGPGSKML 1151
> SPACUNK4.12c
Length=969
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 76/337 (22%), Positives = 136/337 (40%), Gaps = 23/337 (6%)
Query 117 IPKASELEILPGLLGLNEPELISEQGGNAGTAVWWQGQGFSALPR--VAVQLSGSIAKDK 174
IP + E+E P L P L+ + +W + +P+ V + IA+
Sbjct 492 IPWSLEVEKQPVTTKLKVPNLVR---NDKFVRLWHKKDDTFWVPKANVFINFISPIARRS 548
Query 175 ADLLSRTQGSVALAAIAEHLQEETADFQY----CGITHSLAFKGTGFHMTFEGYTKTQLD 230
+ SV+ +++ ++ Y G++ SL+ G + G+T +L
Sbjct 549 PKV------SVSTTLYTRLIEDALGEYSYPASLAGLSFSLSPSTRGIILCISGFTD-KLH 601
Query 231 KLMSHVASVLRDPSVVESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRM 290
L+ V +++RD V +RFE +K + + L D A+ + L+ ++S
Sbjct 602 VLLEKVVAMMRDLKV-HPQRFEILKNRLEQELKDYDALEAYHRSNHVLTWLSEPHSWSNA 660
Query 291 DLLNALEQTTYDDSIAKLSEL-KNVHVDAFVMGNIDRDQSLLLTESFLEQAGFTPIEHDD 349
+L A++ D +S+L K +++ V GN + + L ES + P+
Sbjct 661 ELREAIKDVQVGDMSDFISDLLKQNFLESLVHGNYTEEDAKNLIESAQKLIDPKPVFASQ 720
Query 350 AVESLAM---EQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILS-QFLNRR 405
A+ E I T+ P K +K+ A + Q+ E L L+ Q +
Sbjct 721 LSRKRAIIVPEGGNYIYKTVV-PNKEEKNSAIMYNLQISQLDDERSGALTRLARQIMKEP 779
Query 406 IYDSLRTEAQLGYIAGAKESQAASTALLQCFVEGSKS 442
+ LRT+ QLGYI Q L FV+ +S
Sbjct 780 TFSILRTKEQLGYIVFTLVRQVTPFINLNIFVQSERS 816
> CE01818
Length=745
Score = 45.1 bits (105), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 63/275 (22%), Positives = 109/275 (39%), Gaps = 15/275 (5%)
Query 267 TNMAFE--HALQAAAI--LTRNDAFSRMDLLNALEQTTYDDSIA-KLSELKNVHVDAFVM 321
TN AF H L A I L ++ +S+ LL + T +D + L+ H++ FV
Sbjct 460 TNHAFSQPHDLSAHFIDLLVVDNIWSKEQLLAVCDSVTLEDVHGFAIKMLQAFHMELFVH 519
Query 322 GNIDRDQSLLLTESFLE-----QAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDH 376
GN +L L++ + P++ D+ ++ E + K
Sbjct 520 GNSTEKDTLQLSKELSDILKSVAPNSRPLKRDEHNPHRELQLINGHEHVYRHFQKTHDVG 579
Query 377 ASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCF 436
V FQ+GV S +L++ + Y LRT LGY + L
Sbjct 580 CVEVAFQIGVQSTYNNSVNKLLNELIKNPAYTILRTNEALGYNVSTESRLNDGNVYLHVI 639
Query 437 VEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSA 496
V+G +S D V++ I+ L A++ I MP+ + A + S+ F
Sbjct 640 VQGPES-ADHVLERIEVFLESAREEIVAMPQEDFDY---QVWAMFKENPPTLSQCFSMFW 695
Query 497 DEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTK 531
EI S F R+ EV+ + +++ ++ F +
Sbjct 696 SEIHSRQYNFG-RNKEVRGISKRITKEEVINFFDR 729
> 7296373
Length=908
Score = 45.1 bits (105), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 59/321 (18%), Positives = 137/321 (42%), Gaps = 17/321 (5%)
Query 205 GITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLR--DPSVVESERFERVKQKQIK-- 260
G+T+ L G M GY + +L L+ + ++++ + + + F+ +K++QI
Sbjct 484 GLTYGLYIGDKGLVMRVSGYNE-KLPLLVEIILNMMQTIELDIGQVNAFKDLKKRQIYNA 542
Query 261 LLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE--LKNVHVDA 318
L+ + N+ ++ N FS + +++ T DD I E K ++V
Sbjct 543 LINGKSLNLDLRLSI------LENKRFSMISKYESVDDITMDD-IKSFKENFHKKMYVKG 595
Query 319 FVMGNIDRDQSLLLTESFLEQAGFTPIEHDDAVESLAMEQKQTIEATLANPIKGDKDHAS 378
+ GN DQ+ L + L+ +++ A+++ ++ A + D +
Sbjct 596 LIQGNFTEDQATDLMQKVLDTYKSEKLDNLSALDNHLLQIPLGSYYLRAKTLNEDDSNTI 655
Query 379 LVQF-QLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKE--SQAASTALLQC 435
+ + Q+G ++ + ++ + ++ LRT+ QLGY G + L+
Sbjct 656 ITNYYQIGPSDLKMECIMDLVELIVEEPFFNQLRTQEQLGYSLGIHQRIGYGVLAFLITI 715
Query 436 FVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKS 495
+ +K D V + I+A ++ + +S M + E +E + + E+ ++
Sbjct 716 NTQETKHRADYVEQRIEAFRSRMAELVSQMSDTEFKNIRETLINGKKLGDTSLDEEVLRN 775
Query 496 ADEIFSHANCFSKRDLEVKYL 516
EI + F++ + +++ L
Sbjct 776 WSEIVTKEYFFNRIETQIQML 796
> At2g41790
Length=970
Score = 44.7 bits (104), Expect = 7e-04, Method: Compositional matrix adjust.
Identities = 67/347 (19%), Positives = 139/347 (40%), Gaps = 33/347 (9%)
Query 190 IAEHLQEETADFQYCGITHSLAFKGTGFHMTFEGYT---KTQLDKLMSHVASVLRDPSVV 246
+ ++L E Q G+ + ++ GF +T GY + L+ ++ +A+ P
Sbjct 560 LMDYLNEYAYYAQVAGLYYGVSLSDNGFELTLLGYNHKLRILLETVVGKIANFEVKP--- 616
Query 247 ESERFERVKQKQIKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIA 306
+RF +K+ K + + A+ +++ ++ + + L+ L +D +A
Sbjct 617 --DRFAVIKETVTKEYQNYKFRQPYHQAMYYCSLILQDQTWPWTEELDVLSHLEAED-VA 673
Query 307 KLSE--LKNVHVDAFVMGNIDRDQSLLLTESFLEQAGFT----------PIEH-DDAVES 353
K L ++ ++ GN++ +++ + + +E F P +H + V
Sbjct 674 KFVPMLLSRTFIECYIAGNVENNEAESMVKH-IEDVLFNDPKPICRPLFPSQHLTNRVVK 732
Query 354 LAMEQKQTIEATLANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTE 413
L K +NP D++ A + Q+ + L + + + LRT
Sbjct 733 LGEGMKYFYHQDGSNP--SDENSALVHYIQVHRDDFSMNIKLQLFGLVAKQATFHQLRTV 790
Query 414 AQLGYIAGAKESQAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAELSRW 473
QLGYI + + +Q ++ S P + +++ L K++ S + E +
Sbjct 791 EQLGYITALAQRNDSGIYGVQFIIQSSVKGPGHIDSRVESLL---KNFESKLYEMSNEDF 847
Query 474 KEAAHA----KLTKVEANFSEDFKKSADEIFSHANCFSKRDLEVKYL 516
K A KL K N E+ + EI S F++++ EV L
Sbjct 848 KSNVTALIDMKLEK-HKNLKEESRFYWREIQSGTLKFNRKEAEVSAL 893
> At1g06900
Length=1023
Score = 42.4 bits (98), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 83/422 (19%), Positives = 174/422 (41%), Gaps = 52/422 (12%)
Query 150 WWQGQGFSALPRVA----VQLSGSIAKDKADLLSRTQGSVALAAIAEHLQEETADFQYCG 205
W++ +PR + L G+ A K LL+ + + + + L E +Q
Sbjct 589 WYKLDETFKVPRANTYFRINLKGAYASVKNCLLTE----LYINLLKDELNEII--YQATK 642
Query 206 ITHSLAFKGTGFHMTFEGYTKTQLDKLMSHVASVLRD--PSVVESERFERVKQKQIKLLA 263
+ SL+ G + G+ + ++ L+S + ++ + P++ ERF+ +K+ +
Sbjct 643 LETSLSMYGDKLELKVYGFNE-KIPALLSKILAIAKSFMPNL---ERFKVIKENMERGFR 698
Query 264 DPATNMA-FEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSELKN-VHVDAFVM 321
+ TNM H+ L + + L+ L + DD + + EL++ + ++A
Sbjct 699 N--TNMKPLNHSTYLRLQLLCKRIYDSDEKLSVLNDLSLDDLNSFIPELRSQIFIEALCH 756
Query 322 GNIDRDQSLLLTESFLEQAGFTPI----EHDDAVESLAMEQKQTIEATLANPIKGDKDHA 377
GN+ D+++ ++ F + P+ H + + M K + + N K + +
Sbjct 757 GNLSEDEAVNISNIFKDSLTVEPLPSKCRHGEQITCFPMGAKLVRDVNVKN--KSETNSV 814
Query 378 SLVQFQLG---VPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQ 434
+ +Q+ S + L + + + +++ LRT+ QLGY+
Sbjct 815 VELYYQIEPEEAQSTRTKAVLDLFHEIIEEPLFNQLRTKEQLGYVVECGPRLTYRVHGFC 874
Query 435 CFVEGSKSHP-------DEVVKMIDAELAK-----AKDYISNMPEAELSRWKEAAHAKLT 482
V+ SK P D +K I+ L + +DY S M ++R E + L+
Sbjct 875 FCVQSSKYGPVHLLGRVDNFIKDIEGLLEQLDDESYEDYRSGM----IARLLEKDPSLLS 930
Query 483 KVEANFSEDFKKSADEIFSHANCFSKRDLEVKYLDNDFSRKHLLRTFTKLSDP-SRRMVV 541
+ +S+ K FSH R ++ K + + + +T+ + S P RR+ V
Sbjct 931 ETNDLWSQIVDKRYMFDFSHKEAEELRSIQKKDVISWY------KTYFRESSPKCRRLAV 984
Query 542 KL 543
++
Sbjct 985 RV 986
> 7292708
Length=1077
Score = 41.6 bits (96), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 31/130 (23%), Positives = 58/130 (44%), Gaps = 2/130 (1%)
Query 382 FQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKESQAASTALLQCFVEG-- 439
+Q+G ++ L +L F++ ++D LRT+ QLGY GA A V
Sbjct 814 YQIGPNTVRVESILDLLMMFVDEPLFDQLRTKEQLGYHVGATVRLNYGIAGYSIMVNSQE 873
Query 440 SKSHPDEVVKMIDAELAKAKDYISNMPEAELSRWKEAAHAKLTKVEANFSEDFKKSADEI 499
+K+ D V I+ AK + ++P+ E +++ + S + ++ DEI
Sbjct 874 TKTTADYVEGRIEVFRAKMLQILRHLPQDEYEHTRDSLIKLKLVADLALSTEMSRNWDEI 933
Query 500 FSHANCFSKR 509
+ + F +R
Sbjct 934 INESYLFDRR 943
> At3g57470
Length=989
Score = 39.3 bits (90), Expect = 0.024, Method: Compositional matrix adjust.
Identities = 54/285 (18%), Positives = 109/285 (38%), Gaps = 25/285 (8%)
Query 202 QYCGITHSLAFKGTGFHMTFEGYT---KTQLDKLMSHVASVLRDPSVVESERFERVKQKQ 258
Q G+ + L+ GF ++ G+ + L+ ++ +A P +RF +K+
Sbjct 597 QAAGLDYGLSLSDNGFELSLAGFNHKLRILLEAVIQKIAKFEVKP-----DRFSVIKETV 651
Query 259 IKLLADPATNMAFEHALQAAAILTRNDAFSRMDLLNALEQTTYDDSIAKLSE--LKNVHV 316
K + E A +++ ++ + + L+AL +D +A L V
Sbjct 652 TKAYQNNKFQQPHEQATNYCSLVLQDQIWPWTEELDALSHLEAED-LANFVPMLLSRTFV 710
Query 317 DAFVMGNIDRDQSLLLTESFLEQAGFT---PIEH--------DDAVESLAMEQKQTIEAT 365
+ ++ GN+++D++ + + +E FT PI + V L K
Sbjct 711 ECYIAGNVEKDEAESMVKH-IEDVLFTDSKPICRPLFPSQFLTNRVTELGTGMKHFYYQE 769
Query 366 LANPIKGDKDHASLVQFQLGVPSIEERVNLAILSQFLNRRIYDSLRTEAQLGYIAGAKES 425
+N D++ A + Q+ L + + + LRT QLGYI S
Sbjct 770 GSN--SSDENSALVHYIQVHKDEFSMNSKLQLFELIAKQDTFHQLRTIEQLGYITSLSLS 827
Query 426 QAASTALLQCFVEGSKSHPDEVVKMIDAELAKAKDYISNMPEAEL 470
+ +Q ++ S P + +++ L + NM + E
Sbjct 828 NDSGVYGVQFIIQSSVKGPGHIDSRVESLLKDLESKFYNMSDEEF 872
> At5g67320
Length=613
Score = 34.3 bits (77), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 14/42 (33%), Positives = 27/42 (64%), Gaps = 0/42 (0%)
Query 63 LLSTRRRNKTKNSRERERERERERERERERERERERERERER 104
+L ++R + +ER+R +E ++ ERE E +R R +E++R
Sbjct 97 MLREKKRKERDMEKERDRSKENDKGVEREHEGDRNRAKEKDR 138
Score = 31.6 bits (70), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 13/32 (40%), Positives = 22/32 (68%), Gaps = 0/32 (0%)
Query 76 RERERERERERERERERERERERERERERPNA 107
+ +ER+ E+ER+R +E ++ ERE E +R A
Sbjct 102 KRKERDMEKERDRSKENDKGVEREHEGDRNRA 133
> 7292130
Length=1430
Score = 32.0 bits (71), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 15/29 (51%), Positives = 24/29 (82%), Gaps = 0/29 (0%)
Query 76 RERERERERERERERERERERERERERER 104
+ER+R +ER+R ER+R +ER+R +ER+R
Sbjct 442 KERQRSKERQRSIERQRSKERQRSKERQR 470
> Hs4503509
Length=1382
Score = 31.2 bits (69), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 20/48 (41%), Positives = 32/48 (66%), Gaps = 8/48 (16%)
Query 72 TKNSRERERERERERERE--------RERERERERERERERPNAFRMP 111
++ S ERE +RE+ER+R+ ++ ERER+RER+ +R + FR P
Sbjct 1195 SRPSEEREWDREKERDRDNQDREENDKDPERERDRERDVDREDRFRRP 1242
> At5g16780
Length=820
Score = 30.8 bits (68), Expect = 8.7, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 25/31 (80%), Gaps = 0/31 (0%)
Query 76 RERERERERERERERERERERERERERERPN 106
RE+++ R+R +E+++E+ER R ++RE ER N
Sbjct 86 REKDKSRDRVKEKDKEKERNRHKDRENERDN 116
Lambda K H
0.316 0.132 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 17533460488
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40