bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0480_orf1
Length=219
Score E
Sequences producing significant alignments: (Bits) Value
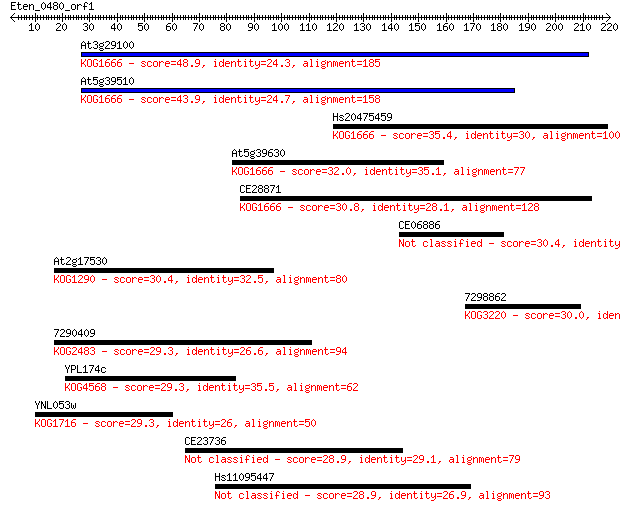
At3g29100 48.9 7e-06
At5g39510 43.9 3e-04
Hs20475459 35.4 0.083
At5g39630 32.0 1.0
CE28871 30.8 2.3
CE06886 30.4 2.6
At2g17530 30.4 3.0
7298862 30.0 4.3
7290409 29.3 5.8
YPL174c 29.3 5.9
YNL053w 29.3 7.0
CE23736 28.9 8.8
Hs11095447 28.9 9.1
> At3g29100
Length=221
Score = 48.9 bits (115), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 45/192 (23%), Positives = 85/192 (44%), Gaps = 20/192 (10%)
Query 27 QLKLRMQNAQSALKRAELTPRS--PHVAAALATALSDADVTLQQLSIEARGSPSGQALAA 84
++K ++ A++ +K+ +L R+ P+V ++L L + L E + SG A
Sbjct 40 EIKSGVEEAEALVKKMDLEARNLPPNVKSSLLVKLREYKSDLNNFKTEVKRITSGNLNAT 99
Query 85 ---EL--AAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELAEVSSRLILESNSSALQ 139
EL A D L A A + R + + L + R I +S + L+
Sbjct 100 ARDELLEAGMADTLT------------ASADQRSRLMMSTDHLGRTTDR-IKDSRRTILE 146
Query 140 TEDIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLNRLLLTGTILL 199
TE++G+ I+Q L QR ++++ + G + ++ + M R N+ + I +
Sbjct 147 TEELGVSILQDLHGQRQSLLRAHETLHGVDDNVGKSKKILTTMTRRMNRNKWTIGAIITV 206
Query 200 LALCILLVLLHK 211
L L I+ +L K
Sbjct 207 LVLAIIFILYFK 218
> At5g39510
Length=221
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 39/163 (23%), Positives = 75/163 (46%), Gaps = 16/163 (9%)
Query 27 QLKLRMQNAQSALKRAELTPRS--PHVAAALATALSDADVTLQQLSIEARGSPSGQALAA 84
++K ++NA+ +++ +L R+ P++ ++L L + L E + SGQ AA
Sbjct 40 EIKSGLENAEVLIRKMDLEARTLPPNLKSSLLVKLREFKSDLNNFKTEVKRITSGQLNAA 99
Query 85 ---ELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELAEVSSRLILESNSSALQTE 141
EL G A T A A + R + E L + R + +S + ++TE
Sbjct 100 ARDELLEAGMA----------DTKTASADQRARLMMSTERLGRTTDR-VKDSRRTMMETE 148
Query 142 DIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIR 184
+IG+ I+Q L QR ++++ + G + ++ + M R
Sbjct 149 EIGVSILQDLHGQRQSLLRAHETLHGVDDNIGKSKKILTDMTR 191
> Hs20475459
Length=231
Score = 35.4 bits (80), Expect = 0.083, Method: Compositional matrix adjust.
Identities = 30/100 (30%), Positives = 52/100 (52%), Gaps = 6/100 (6%)
Query 119 PEELAEVSSRLILESNSSALQTEDIGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQA 178
PE L +++ I S+ A +T+ IG +++L QRD + +T T ++ R+
Sbjct 135 PENLNR-ATQSIERSHQIATETDQIGSETIEELGEQRDHLERTKSRLINTTENLSKSRKI 193
Query 179 IVKMIRDHWLNRLLLTGTILLLALCILLVLLHKARLFRFF 218
+ M R N+LL + I+LL L +L L++ +RFF
Sbjct 194 LRSMSRKVTTNKLLFS-IIILLELAVLGGLVY----YRFF 228
> At5g39630
Length=207
Score = 32.0 bits (71), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 46/92 (50%), Gaps = 15/92 (16%)
Query 82 LAAELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPE--ELAEVSSRL---------- 129
L +L +L RLR + + TS + T + LE E +LA+ SRL
Sbjct 70 LLEKLKESKSSLKRLRNEIKRNTSENLKVTTREEVLEAEKADLADQRSRLMKSTEGLVRT 129
Query 130 ---ILESNSSALQTEDIGLHIMQQLRTQRDAI 158
I +S L+TE+IG+ I++ L+ Q++++
Sbjct 130 REMIKDSQRKLLETENIGISILENLQRQKESL 161
> CE28871
Length=224
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 62/128 (48%), Gaps = 15/128 (11%)
Query 85 ELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELAEVSSRLILESNSSALQTEDIG 144
EL A+ D L RQ+ + + T R E S+R + +++ A++TE IG
Sbjct 111 ELLAFDDQLDEHRQEDQ------LIANTQR--------LERSTRKVQDAHRIAVETEQIG 156
Query 145 LHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIVKMIRDHWLNRLLLTGTILLLALCI 204
++ L +QR+ I ++ + A+ R + + MIR NRLLL LL+ +
Sbjct 157 AEMLSNLASQRETIGRSRDRLRQSNADLGRANKTLSSMIRRAIQNRLLLLIVTFLLSF-M 215
Query 205 LLVLLHKA 212
L +++KA
Sbjct 216 FLYIVYKA 223
> CE06886
Length=345
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 16/38 (42%), Positives = 22/38 (57%), Gaps = 0/38 (0%)
Query 143 IGLHIMQQLRTQRDAIIKTNRLAEGTGAEGARGRQAIV 180
+GL ++QL ++R A I T L TG EGAR I+
Sbjct 296 VGLISLRQLNSRRHAKILTKVLIRVTGQEGARNYDDII 333
> At2g17530
Length=440
Score = 30.4 bits (67), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 38/80 (47%), Gaps = 7/80 (8%)
Query 17 KTTNYRALEQQLKLRMQNAQSALKRAELTPRSPHVAAALATALSDADVTLQQLSIEARGS 76
+T+NY AL+ Q K +Q AQ+AL EL + AA + ++ +
Sbjct 59 RTSNYVALKIQ-KSALQFAQAALHEIEL------LQAAADGDPENTKCVIRLIDDFKHAG 111
Query 77 PSGQALAAELAAYGDALARL 96
P+GQ L L GD+L RL
Sbjct 112 PNGQHLCMVLEFLGDSLLRL 131
> 7298862
Length=236
Score = 30.0 bits (66), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 14/45 (31%), Positives = 23/45 (51%), Gaps = 3/45 (6%)
Query 167 GTGAEGARGRQAIVKMIRD---HWLNRLLLTGTILLLALCILLVL 208
G+ E +I ++RD HWLNR+ G L++ I ++L
Sbjct 180 GSVEEAESSAMSIYNLMRDSKQHWLNRISFLGLFLIVGFTIYMLL 224
> 7290409
Length=706
Score = 29.3 bits (64), Expect = 5.8, Method: Composition-based stats.
Identities = 25/94 (26%), Positives = 42/94 (44%), Gaps = 2/94 (2%)
Query 17 KTTNYRALEQQLKLRMQNAQSALKRAELTPRSPHVAAALATALSDADVTLQQLSIEARGS 76
K + +E+Q ++ ++N ALK+ T R A + A + +Q E S
Sbjct 615 KRNQHNDMERQRRIGLKNLFEALKKQIPTIRDKERAPKVNILREAAKLCIQLTQEEKELS 674
Query 77 PSGQALAAELAAYGDALARLRQQSEEWTSRAVAG 110
Q L+ +L D LA + + E SR+V+G
Sbjct 675 MQRQLLSLQLKQRQDTLASYQMELNE--SRSVSG 706
> YPL174c
Length=868
Score = 29.3 bits (64), Expect = 5.9, Method: Composition-based stats.
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 8/62 (12%)
Query 21 YRALEQQLKLRMQNAQSALKRAELTPRSPHVAAALATALSDADVTLQQLSIEARGSPSGQ 80
Y LEQ+L+L++ N QSAL+ + +A S TL+ SIEA+ S Q
Sbjct 311 YEQLEQELRLQLSNLQSALENEK------EIAGTYIEENSRLKATLE--SIEAKTSHKFQ 362
Query 81 AL 82
+L
Sbjct 363 SL 364
> YNL053w
Length=489
Score = 29.3 bits (64), Expect = 7.0, Method: Compositional matrix adjust.
Identities = 13/50 (26%), Positives = 25/50 (50%), Gaps = 0/50 (0%)
Query 10 AAATAAYKTTNYRALEQQLKLRMQNAQSALKRAELTPRSPHVAAALATAL 59
++ T +Y + ++R+ L S++ +E+TPR+P TAL
Sbjct 399 SSTTKSYSSASFRSFPMVTNLSSSPNDSSVNSSEVTPRTPATLTGARTAL 448
> CE23736
Length=508
Score = 28.9 bits (63), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 7/79 (8%)
Query 65 TLQQLSIEARGSPSGQALAAELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELAE 124
T++++ E G P L +L A DAL +QQ+ + + + LEP E+ E
Sbjct 4 TVEKMETEEYGKPDPNRLTTKLFAEYDALHE-KQQN------GMDEKKTMELLEPSEVFE 56
Query 125 VSSRLILESNSSALQTEDI 143
+ S +E S A + DI
Sbjct 57 IYSPATMEIISIAKEEGDI 75
> Hs11095447
Length=255
Score = 28.9 bits (63), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 25/98 (25%), Positives = 42/98 (42%), Gaps = 5/98 (5%)
Query 76 SPSG--QALAAELAAYGDALARLRQQSEEWTSRAVAGETPRQQLEPEELA---EVSSRLI 130
SP G +A +A+YG + S ++T E LE +E + S+ I
Sbjct 19 SPCGGEDIVADHVASYGVNFYQSHGPSGQYTHEFDGDEEFYVDLETKETVWQLPMFSKFI 78
Query 131 LESNSSALQTEDIGLHIMQQLRTQRDAIIKTNRLAEGT 168
SAL+ +G H ++ + Q ++ TN + E T
Sbjct 79 SFDPQSALRNMAVGKHTLEFMMRQSNSTAATNEVPEVT 116
Lambda K H
0.318 0.128 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4112032690
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40