bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
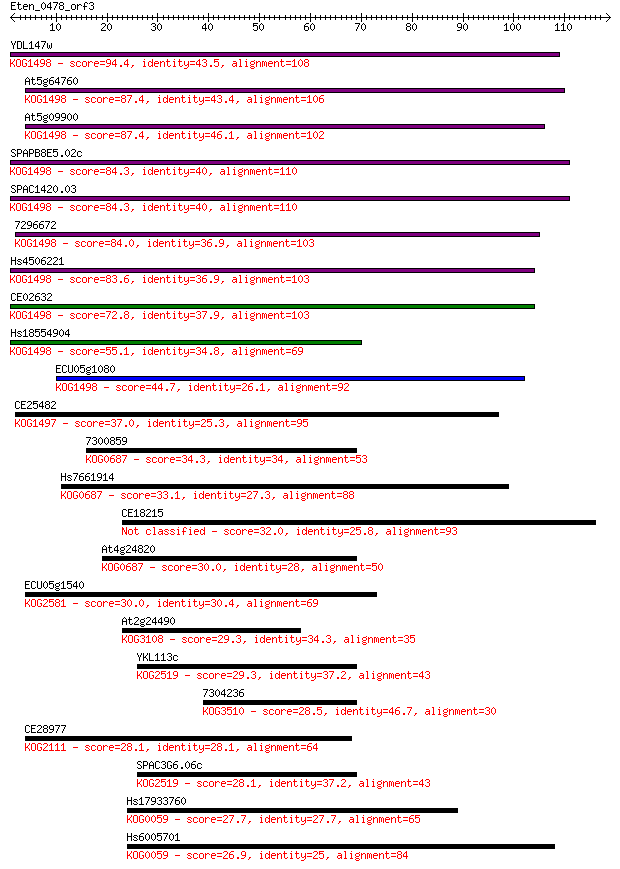
Query= Eten_0478_orf3
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
YDL147w 94.4 5e-20
At5g64760 87.4 5e-18
At5g09900 87.4 6e-18
SPAPB8E5.02c 84.3 5e-17
SPAC1420.03 84.3 5e-17
7296672 84.0 6e-17
Hs4506221 83.6 8e-17
CE02632 72.8 1e-13
Hs18554904 55.1 3e-08
ECU05g1080 44.7 5e-05
CE25482 37.0 0.008
7300859 34.3 0.056
Hs7661914 33.1 0.12
CE18215 32.0 0.25
At4g24820 30.0 1.0
ECU05g1540 30.0 1.0
At2g24490 29.3 1.6
YKL113c 29.3 1.6
7304236 28.5 3.2
CE28977 28.1 3.9
SPAC3G6.06c 28.1 4.4
Hs17933760 27.7 5.1
Hs6005701 26.9 8.1
> YDL147w
Length=445
Score = 94.4 bits (233), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 47/108 (43%), Positives = 73/108 (67%), Gaps = 0/108 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+KRVI+HNLRV+ YS I L R+ LL + E + E+ +S+L + G++YAK++RPA
Sbjct 338 WEDLQKRVIEHNLRVISEYYSRITLLRLNELLDLTESQTETYISDLVNQGIIYAKVNRPA 397
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAK 108
+ V F KPK +L EWS ++ +LL+ ++ H+I KE ++H +AK
Sbjct 398 KIVNFEKPKNSSQLLNEWSHNVDELLEHIETIGHLITKEEIMHGLQAK 445
> At5g64760
Length=529
Score = 87.4 bits (215), Expect = 5e-18, Method: Compositional matrix adjust.
Identities = 46/106 (43%), Positives = 64/106 (60%), Gaps = 0/106 (0%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTV 63
L+ R+I+HN+ V+ YS I +R+A LL + EEAE LSE+ + L AKIDRP+ +
Sbjct 424 LKLRIIEHNILVVSKYYSRITFKRLAELLCLTTEEAEKHLSEMVVSKALIAKIDRPSGII 483
Query 64 AFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAKA 109
F K +L W+++L KLLD V+ CH I KE MVH +A
Sbjct 484 CFQIVKDSNEILNSWATNLEKLLDLVEKSCHQIHKETMVHKVALRA 529
> At5g09900
Length=442
Score = 87.4 bits (215), Expect = 6e-18, Method: Compositional matrix adjust.
Identities = 47/102 (46%), Positives = 63/102 (61%), Gaps = 0/102 (0%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTV 63
L+ R+I+HN+ V+ Y+ I L+R+A LL + EEAE LSE+ + L AKIDRP+ V
Sbjct 337 LKLRIIEHNILVVSKYYARITLKRLAELLCLSMEEAEKHLSEMVVSKALIAKIDRPSGIV 396
Query 64 AFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAA 105
F K +L W+ +L KLLD V+ CH I KE MVH A
Sbjct 397 CFQIAKDSNEILNSWAGNLEKLLDLVEKSCHQIHKETMVHKA 438
> SPAPB8E5.02c
Length=443
Score = 84.3 bits (207), Expect = 5e-17, Method: Composition-based stats.
Identities = 44/110 (40%), Positives = 67/110 (60%), Gaps = 0/110 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W+ LRKRVI+HN+RV+ YS IH R+ LL + E E L +L + YAKIDRPA
Sbjct 333 WSELRKRVIEHNIRVVANYYSRIHCSRLGVLLDMSPSETEQFLCDLIAKHHFYAKIDRPA 392
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAKAA 110
+ ++F K + L EW S++++LL K++ +I KE M+++ + A
Sbjct 393 QVISFKKSQNVQEQLNEWGSNITELLGKLEKVRQLIIKEEMMNSIQQAVA 442
> SPAC1420.03
Length=443
Score = 84.3 bits (207), Expect = 5e-17, Method: Composition-based stats.
Identities = 44/110 (40%), Positives = 67/110 (60%), Gaps = 0/110 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W+ LRKRVI+HN+RV+ YS IH R+ LL + E E L +L + YAKIDRPA
Sbjct 333 WSELRKRVIEHNIRVVANYYSRIHCSRLGVLLDMSPSETEQFLCDLIAKHHFYAKIDRPA 392
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHAARAKAA 110
+ ++F K + L EW S++++LL K++ +I KE M+++ + A
Sbjct 393 QVISFKKSQNVQEQLNEWGSNITELLGKLEKVRQLIIKEEMMNSIQQAVA 442
> 7296672
Length=502
Score = 84.0 bits (206), Expect = 6e-17, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 65/103 (63%), Gaps = 0/103 (0%)
Query 2 TLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPAR 61
T L+ R+I+HN+R++ YS +HL R++ LL +P E LS+LA+ + KIDRPA
Sbjct 393 TELKDRLIEHNIRIIAMYYSRLHLARMSELLNLPTSRCEEYLSKLANTDTIRVKIDRPAG 452
Query 62 TVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVHA 104
+ F + K+ +L W++ +++L+ V+ CH+I KE V++
Sbjct 453 IIYFTQKKSASDILNNWATDVNQLMSLVNKTCHLINKEECVYS 495
> Hs4506221
Length=456
Score = 83.6 bits (205), Expect = 8e-17, Method: Compositional matrix adjust.
Identities = 38/103 (36%), Positives = 64/103 (62%), Gaps = 0/103 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L+ RV++HN+R++ Y+ I ++R+A LL + +E+E+ LS L ++AK+DR A
Sbjct 351 WKDLKNRVVEHNIRIMAKYYTRITMKRMAQLLDLSVDESEAFLSNLVVNKTIFAKVDRLA 410
Query 61 RTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+ F +PK +L +WS L+ L+ V+ H+I KE M+H
Sbjct 411 GIINFQRPKDPNNLLNDWSQKLNSLMSLVNKTTHLIAKEEMIH 453
> CE02632
Length=490
Score = 72.8 bits (177), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 39/104 (37%), Positives = 62/104 (59%), Gaps = 1/104 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAG-VLYAKIDRP 59
W+ L RV +HN+R++ Y+ I +R+A LL P +E ES + L G + AK+ RP
Sbjct 377 WSDLHLRVGEHNMRMIAKYYTQITFERLAELLDFPVDEMESFVCNLIVTGQITGAKLHRP 436
Query 60 ARTVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMIQKERMVH 103
+R V K ++ L+ W+S++ KL D ++ H+I KE+MVH
Sbjct 437 SRIVNLRLKKANVEQLDVWASNVHKLTDTLNKVSHLILKEQMVH 480
> Hs18554904
Length=113
Score = 55.1 bits (131), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 24/69 (34%), Positives = 42/69 (60%), Gaps = 0/69 (0%)
Query 1 WTLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPA 60
W L RV++HN+R++ Y+ + ++R+ LL + +E+E+ LS L ++AK+DR A
Sbjct 40 WKDLNNRVVEHNIRIMAKYYTRMTMKRMVQLLDLSVDESEAFLSNLVVNKTIFAKVDRLA 99
Query 61 RTVAFGKPK 69
F +PK
Sbjct 100 GINNFQRPK 108
> ECU05g1080
Length=387
Score = 44.7 bits (104), Expect = 5e-05, Method: Composition-based stats.
Identities = 24/92 (26%), Positives = 48/92 (52%), Gaps = 8/92 (8%)
Query 10 QHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPK 69
+HN R++ YS I +Q ++ ++ P E+ ++S + + G KI++ + F K K
Sbjct 297 EHNFRIIERFYSSISVQEISMVMQSPAEDIIKKISFMVNNGFAQCKINQKTGIIEFEKRK 356
Query 70 TDLVVLEEWSSSLSKLLDKVDLCCHMIQKERM 101
W+ ++ ++ K+ C H+I KER+
Sbjct 357 --------WNDNVEDVMGKLIKCNHLIHKERL 380
> CE25482
Length=412
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 4/95 (4%)
Query 2 TLLRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPAR 61
++L+ + +HN+ + Y I + + LLG+ E AES E+ S+ L+ ID+
Sbjct 310 SILKGVIQEHNITAISQLYINISFKTLGQLLGVDTEAAESMAGEMISSERLHGYIDQTNG 369
Query 62 TVAFGKPKTDLVVLEEWSSSLSKLLDKVDLCCHMI 96
+ F D + W S + L++++ MI
Sbjct 370 ILHF----EDSNPMRVWDSQILSTLEQINKVSDMI 400
> 7300859
Length=389
Score = 34.3 bits (77), Expect = 0.056, Method: Composition-based stats.
Identities = 18/53 (33%), Positives = 28/53 (52%), Gaps = 0/53 (0%)
Query 16 LCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
L +Y + LQ +A G+ E + EL+ +AG L+AK+DR V +P
Sbjct 307 LLESYRSLTLQYMAESFGVTVEYIDQELARFIAAGRLHAKVDRVGGIVETNRP 359
> Hs7661914
Length=389
Score = 33.1 bits (74), Expect = 0.12, Method: Compositional matrix adjust.
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 7/88 (7%)
Query 11 HNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKT 70
H L +Y + L +A G+ E + ELS +AG L+ KID+ V +P +
Sbjct 302 HAYSQLLESYRSLTLGYMAEAFGVGVEFIDQELSRFIAAGRLHCKIDKVNEIVETNRPDS 361
Query 71 DLVVLEEWSSSLSKLLDKVDLCCHMIQK 98
+ W + + K DL + +QK
Sbjct 362 -----KNW--QYQETIKKGDLLLNRVQK 382
> CE18215
Length=275
Score = 32.0 bits (71), Expect = 0.25, Method: Compositional matrix adjust.
Identities = 24/109 (22%), Positives = 44/109 (40%), Gaps = 17/109 (15%)
Query 23 IHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSS-- 80
+HL+++ + GI EE ++ L L + + D P D+V LE+W +
Sbjct 114 LHLEKI-NFKGINYEEIDTILRHLKTGVLKEISFDYKITNSTIIPPIGDMVYLEQWKNAE 172
Query 81 --------SLSKLLD------KVDLCCHMIQKERMVHAARAKAAAANAR 115
SL LLD +D+ CH + +++ + + R
Sbjct 173 NLCIHSHISLGSLLDHISHFHTIDILCHTFSRLQILQLQKIIDTSKTMR 221
> At4g24820
Length=406
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 14/50 (28%), Positives = 27/50 (54%), Gaps = 0/50 (0%)
Query 19 NYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
+Y + ++ +A G+ + + ELS +AG L+ KID+ A + +P
Sbjct 327 SYKSVTVEAMAKAFGVSVDFIDQELSRFIAAGKLHCKIDKVAGVLETNRP 376
> ECU05g1540
Length=376
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 34/70 (48%), Gaps = 1/70 (1%)
Query 4 LRKRVIQHNLRVLCANYSVIHLQRVASLLGIPEEEAESELSELASAGVLYAKI-DRPART 62
L + VIQ +R + YS I + +A +LGI E E + G++ K+ D +
Sbjct 270 LSQNVIQEGIRKISVVYSRISYEDIAHILGINSGEVEYLVKRTIRKGLIKGKVADGIFYS 329
Query 63 VAFGKPKTDL 72
+ K KTD+
Sbjct 330 LREDKSKTDI 339
> At2g24490
Length=250
Score = 29.3 bits (64), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 20/35 (57%), Gaps = 0/35 (0%)
Query 23 IHLQRVASLLGIPEEEAESELSELASAGVLYAKID 57
IH+ +A L IP+ + E + L G++Y+ ID
Sbjct 207 IHIDEIAQQLKIPKNKLEGVVQSLEGDGLIYSTID 241
> YKL113c
Length=382
Score = 29.3 bits (64), Expect = 1.6, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 26 QRVASLLGIP----EEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
Q++ L+GIP EAE++ +ELA G +YA T+ + P
Sbjct 140 QKLLGLMGIPYIIAPTEAEAQCAELAKKGKVYAAASEDMDTLCYRTP 186
> 7304236
Length=2016
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 39 AESELSELASAGVLYAKIDRPARTVAFGKP 68
ES + L L AKID P +TV FG+P
Sbjct 330 GESVETVLTVTAPLSAKIDPPTQTVDFGRP 359
> CE28977
Length=440
Score = 28.1 bits (61), Expect = 3.9, Method: Composition-based stats.
Identities = 18/73 (24%), Positives = 38/73 (52%), Gaps = 9/73 (12%)
Query 4 LRKRVIQHNLRVL----CANY-SVIHLQRVASLLGIPEEEAESELSEL----ASAGVLYA 54
LR+ +Q +L+ + C++Y +V + + GI + E + + + L S+ ++
Sbjct 312 LRRGTVQAHLQCMAFSPCSSYLAVASDKGTLHMFGIRDAEPQKKKNVLERSRGSSSIVKI 371
Query 55 KIDRPARTVAFGK 67
++DRP + FGK
Sbjct 372 QLDRPVMAIGFGK 384
> SPAC3G6.06c
Length=380
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 26 QRVASLLGIP----EEEAESELSELASAGVLYAKIDRPARTVAFGKP 68
+R+ L+GIP EAE++ + LA +G +YA T+ F P
Sbjct 143 KRLLELMGIPFVNAPCEAEAQCAALARSGKVYAAASEDMDTLCFQAP 189
> Hs17933760
Length=1543
Score = 27.7 bits (60), Expect = 5.1, Method: Composition-based stats.
Identities = 18/65 (27%), Positives = 33/65 (50%), Gaps = 0/65 (0%)
Query 24 HLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSSLS 83
HL+ A++ G+ +E+A +S L A L ++ P +T++ G + VL +
Sbjct 1301 HLELYAAVKGLGKEDAALSISRLVEALKLQEQLKAPVKTLSEGIKRKLCFVLSILGNPSV 1360
Query 84 KLLDK 88
LLD+
Sbjct 1361 VLLDE 1365
> Hs6005701
Length=1581
Score = 26.9 bits (58), Expect = 8.1, Method: Composition-based stats.
Identities = 21/84 (25%), Positives = 40/84 (47%), Gaps = 0/84 (0%)
Query 24 HLQRVASLLGIPEEEAESELSELASAGVLYAKIDRPARTVAFGKPKTDLVVLEEWSSSLS 83
HL+ A++ G+ + +AE ++ L A L ++ P +T++ G + VL +
Sbjct 1339 HLEVYAAVKGLRKGDAEVAITRLVDALKLQDQLKSPVKTLSEGIKRKLCFVLSILGNPSV 1398
Query 84 KLLDKVDLCCHMIQKERMVHAARA 107
LLD+ +++M A RA
Sbjct 1399 VLLDEPSTGMDPEGQQQMWQAIRA 1422
Lambda K H
0.320 0.129 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167969826
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40