bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0436_orf2
Length=178
Score E
Sequences producing significant alignments: (Bits) Value
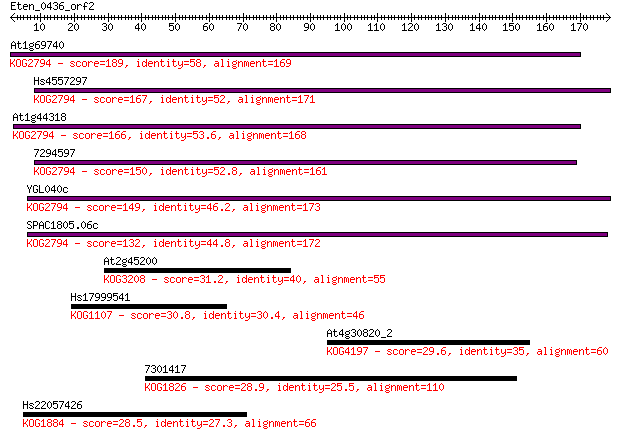
At1g69740 189 2e-48
Hs4557297 167 1e-41
At1g44318 166 2e-41
7294597 150 2e-36
YGL040c 149 2e-36
SPAC1805.06c 132 4e-31
At2g45200 31.2 1.3
Hs17999541 30.8 1.5
At4g30820_2 29.6 3.4
7301417 28.9 6.2
Hs22057426 28.5 6.7
> At1g69740
Length=430
Score = 189 bits (480), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 98/169 (57%), Positives = 124/169 (73%), Gaps = 1/169 (0%)
Query 1 CKQPFNLARAGADMVCPSEMMDGRVTAIREALDMEGCVDTSVLSYACKYASSLYGPFRDA 60
CKQ + ARAGAD+V PS+MMDGRV AIR ALD EG + S++SY KYASS YGPFR+A
Sbjct 251 CKQAVSQARAGADVVSPSDMMDGRVGAIRSALDAEGFQNVSIMSYTAKYASSFYGPFREA 310
Query 61 VGSPLKGTGDKKTYQMDISNALEAEREAELDVQEGADMLMVKPGSSYLDVLRRVRQKTNV 120
+ S + GDKKTYQM+ +N EA EA D EGAD+L+VKPG YLD++R +R K+ +
Sbjct 311 LDSNPR-FGDKKTYQMNPANYREALIEAREDEAEGADILLVKPGLPYLDIIRLLRDKSPL 369
Query 121 PLAVYQVSGEYAMIKAAAEKGWINEKAVVLETLKGFRRAGADAIATYYA 169
P+A YQVSGEY+MIKA I+E+ V++E+L RRAGAD I TY+A
Sbjct 370 PIAAYQVSGEYSMIKAGGVLKMIDEEKVMMESLMCLRRAGADIILTYFA 418
> Hs4557297
Length=330
Score = 167 bits (422), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 89/173 (51%), Positives = 115/173 (66%), Gaps = 3/173 (1%)
Query 8 ARAGADMVCPSEMMDGRVTAIREALDMEGCVD-TSVLSYACKYASSLYGPFRDAVGSPLK 66
A+AG +V PS+MMDGRV AI+EAL G + SV+SY+ K+AS YGPFRDA S
Sbjct 158 AKAGCQVVAPSDMMDGRVEAIKEALMAHGLGNRVSVMSYSAKFASCFYGPFRDAAKSS-P 216
Query 67 GTGDKKTYQMDISNALEAEREAELDVQEGADMLMVKPGSSYLDVLRRVRQK-TNVPLAVY 125
GD++ YQ+ A R + DV+EGADMLMVKPG YLD++R V+ K ++PLAVY
Sbjct 217 AFGDRRCYQLPPGARGLALRAVDRDVREGADMLMVKPGMPYLDIVREVKDKHPDLPLAVY 276
Query 126 QVSGEYAMIKAAAEKGWINEKAVVLETLKGFRRAGADAIATYYAKDFAKWMEE 178
VSGE+AM+ A+ G + KA VLE + FRRAGAD I TYY +W++E
Sbjct 277 HVSGEFAMLWHGAQAGAFDLKAAVLEAMTAFRRAGADIIITYYTPQLLQWLKE 329
> At1g44318
Length=406
Score = 166 bits (421), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 90/168 (53%), Positives = 116/168 (69%), Gaps = 7/168 (4%)
Query 2 KQPFNLARAGADMVCPSEMMDGRVTAIREALDMEGCVDTSVLSYACKYASSLYGPFRDAV 61
KQ + ARAGAD+VC SEM+DGRV A+R ALD EG D S++SY+ KY SSLYG FR
Sbjct 229 KQAVSQARAGADVVCTSEMLDGRVGAVRAALDAEGFQDVSIMSYSVKYTSSLYGRFR--- 285
Query 62 GSPLKGTGDKKTYQMDISNALEAEREAELDVQEGADMLMVKPGSSYLDVLRRVRQKTNVP 121
K DKKTYQ++ +N+ EA EA D EGAD+LMVKP LD++R ++ +T +P
Sbjct 286 ----KVQLDKKTYQINPANSREALLEAREDEAEGADILMVKPALPSLDIIRLLKNQTLLP 341
Query 122 LAVYQVSGEYAMIKAAAEKGWINEKAVVLETLKGFRRAGADAIATYYA 169
+ QVSGEY+MIKAA I+E+ V++E+L RRAGAD I TY+A
Sbjct 342 IGACQVSGEYSMIKAAGLLKMIDEEKVMMESLLCIRRAGADLILTYFA 389
> 7294597
Length=327
Score = 150 bits (378), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 85/164 (51%), Positives = 108/164 (65%), Gaps = 6/164 (3%)
Query 8 ARAGADMVCPSEMMDGRVTAIREALDMEGCVDTSVLSYACKYASSLYGPFRDAVGSPLKG 67
A+AGA +V PS+MMD RV AI++AL S+L+Y+ K+ S+ YGPFR+A S K
Sbjct 155 AKAGAHIVAPSDMMDNRVKAIKQALIDAQMNSVSLLAYSAKFTSNFYGPFREAAQSAPK- 213
Query 68 TGDKKTYQMDISNALEAEREAELDVQEGADMLMVKPGSSYLDVLRRVRQKTNVP---LAV 124
GD++ YQ+ + A R + DV EGADMLMVKPG YLD+LR K + P L V
Sbjct 214 FGDRRCYQLPSGSRSLAMRAIQRDVAEGADMLMVKPGMPYLDILRST--KDSYPYHTLYV 271
Query 125 YQVSGEYAMIKAAAEKGWINEKAVVLETLKGFRRAGADAIATYY 168
YQVSGE+AM+ AA+ G + K VLE +KGFRRAGAD I TYY
Sbjct 272 YQVSGEFAMLYHAAKAGAFDLKDAVLEAMKGFRRAGADCIITYY 315
> YGL040c
Length=342
Score = 149 bits (377), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 80/175 (45%), Positives = 111/175 (63%), Gaps = 3/175 (1%)
Query 6 NLARAGADMVCPSEMMDGRVTAIREAL-DMEGCVDTSVLSYACKYASSLYGPFRDAVGSP 64
N A+AGA V PS+M+DGR+ I+ L + T VLSYA K++ +LYGPFRDA S
Sbjct 167 NYAKAGAHCVAPSDMIDGRIRDIKRGLINANLAHKTFVLSYAAKFSGNLYGPFRDAACS- 225
Query 65 LKGTGDKKTYQMDISNALEAEREAELDVQEGADMLMVKPGSSYLDVLRRVRQKT-NVPLA 123
GD+K YQ+ + A R E D+ EGAD ++VKP + YLD++R + ++P+
Sbjct 226 APSNGDRKCYQLPPAGRGLARRALERDMSEGADGIIVKPSTFYLDIMRDASEICKDLPIC 285
Query 124 VYQVSGEYAMIKAAAEKGWINEKAVVLETLKGFRRAGADAIATYYAKDFAKWMEE 178
Y VSGEYAM+ AAAEKG ++ K + E+ +GF RAGA I TY A +F W++E
Sbjct 286 AYHVSGEYAMLHAAAEKGVVDLKTIAFESHQGFLRAGARLIITYLAPEFLDWLDE 340
> SPAC1805.06c
Length=329
Score = 132 bits (332), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 77/174 (44%), Positives = 110/174 (63%), Gaps = 3/174 (1%)
Query 6 NLARAGADMVCPSEMMDGRVTAIREAL-DMEGCVDTSVLSYACKYASSLYGPFRDAVGSP 64
N A AGA ++ PS+ MDGRV AI++ L ++E V+SY+ K+AS +GPFR A
Sbjct 156 NYALAGAQIISPSDCMDGRVKAIKQKLVELELSHKVCVISYSAKFASGFFGPFRAAANGA 215
Query 65 LKGTGDKKTYQMDISNALEAEREAELDVQEGADMLMVKPGSSYLDVLRRVRQ-KTNVPLA 123
K GD+ YQ+ + A+R DV+EGAD +MVKPG+ YLD+L + ++P+A
Sbjct 216 PK-FGDRSCYQLPCNARGLAKRAILRDVREGADGIMVKPGTPYLDILAMASKLADDLPIA 274
Query 124 VYQVSGEYAMIKAAAEKGWINEKAVVLETLKGFRRAGADAIATYYAKDFAKWME 177
YQVSGE+A+I AAA G K V+ET+ GF RAGA+ + TY+ + +W+E
Sbjct 275 TYQVSGEFAIIHAAAAAGVFELKRHVMETMDGFMRAGANIVLTYFTPELLEWLE 328
> At2g45200
Length=239
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 31/56 (55%), Gaps = 2/56 (3%)
Query 29 REALDMEGCVDTSVLSYACKYASSLYGPFRDAVGSPLKGTGDK-KTYQMDISNALE 83
REA +EG +D + SYA A G + D GSP G+G K+ +M+I + LE
Sbjct 18 REARKIEGDLDVKLSSYAKLGARFTQGGYVD-TGSPTVGSGRSWKSMEMEIQSLLE 72
> Hs17999541
Length=796
Score = 30.8 bits (68), Expect = 1.5, Method: Composition-based stats.
Identities = 14/47 (29%), Positives = 30/47 (63%), Gaps = 1/47 (2%)
Query 19 EMMDGRVTAIR-EALDMEGCVDTSVLSYACKYASSLYGPFRDAVGSP 64
+++D + A++ ++ M+ C+D + L A K+AS++ G R ++ SP
Sbjct 14 KLLDEAIQAVKVQSFQMKRCLDKNKLMDALKHASNMLGELRTSMLSP 60
> At4g30820_2
Length=761
Score = 29.6 bits (65), Expect = 3.4, Method: Composition-based stats.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 10/61 (16%)
Query 95 GADMLMVKPGSSYLDVLRRVRQKTNVP-LAVYQVSGEYAMIKAAAEKGWINEKAVVLETL 153
G D M K + +L+R+++ T+ P Y + MI E+GWI+E A VL+ L
Sbjct 636 GKDKQMEK----FRSILKRMKKSTSGPDHYTYNI-----MINIYGEQGWIDEVADVLKEL 686
Query 154 K 154
K
Sbjct 687 K 687
> 7301417
Length=2802
Score = 28.9 bits (63), Expect = 6.2, Method: Composition-based stats.
Identities = 28/110 (25%), Positives = 49/110 (44%), Gaps = 6/110 (5%)
Query 41 SVLSYACKYASSLYGPFRDAVGSPLKGTGDKKTYQMDISNALEAEREAELDVQEGADMLM 100
+V Y C Y F D + +PLK G++K ++ N L +AE GA + +
Sbjct 1707 AVYIYNCNSWVREYTKFHDRILAPLK--GNRKLLFLESPNKLTDFIDAEQQKLPGATLSL 1764
Query 101 VKPGSSYLDVLRRVRQKTNVPLAVYQVSGEYAMIKAAAEKGWINEKAVVL 150
+ + + L+ + T V + V G A+ +AEK + +V+L
Sbjct 1765 DEDLKVFSNALKLSHKDTKVAIKV----GPTALQITSAEKTKVLAHSVLL 1810
> Hs22057426
Length=2407
Score = 28.5 bits (62), Expect = 6.7, Method: Composition-based stats.
Identities = 18/66 (27%), Positives = 33/66 (50%), Gaps = 4/66 (6%)
Query 5 FNLARAGADMVCPSEMMDGRVTAIREALDMEGCVDTSVLSYACKYASSLYGPFRDAVGSP 64
F++ GA VC E++ + +++ +DM+ V +LSY C+ + Y R++V
Sbjct 1245 FHIPSIGAACVCFLELLG--LDSLKLRVDMK--VANIILSYKCRNEDAQYSFIRESVAEK 1300
Query 65 LKGTGD 70
L D
Sbjct 1301 LSKLAD 1306
Lambda K H
0.317 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 2779358582
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40