bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0409_orf3
Length=106
Score E
Sequences producing significant alignments: (Bits) Value
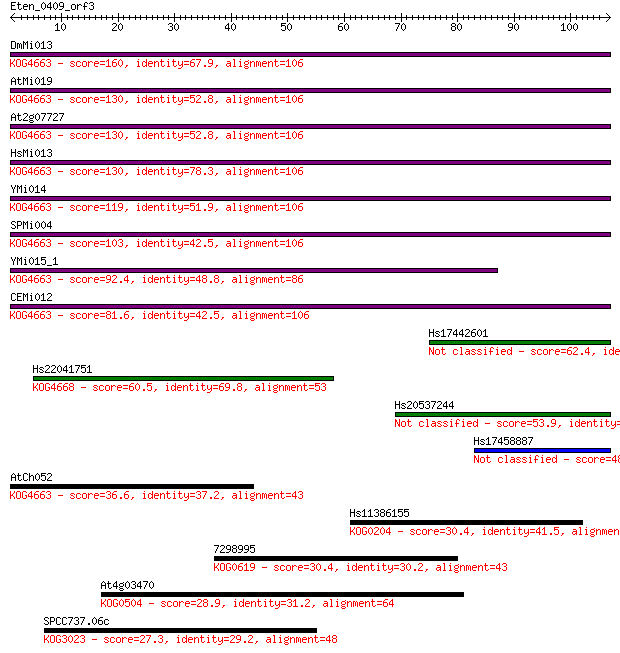
DmMi013 160 7e-40
AtMi019 130 4e-31
At2g07727 130 4e-31
HsMi013 130 4e-31
YMi014 119 2e-27
SPMi004 103 8e-23
YMi015_1 92.4 2e-19
CEMi012 81.6 3e-16
Hs17442601 62.4 2e-10
Hs22041751 60.5 8e-10
Hs20537244 53.9 7e-08
Hs17458887 48.5 3e-06
AtCh052 36.6 0.011
Hs11386155 30.4 0.76
7298995 30.4 0.91
At4g03470 28.9 2.1
SPCC737.06c 27.3 7.5
> DmMi013
Length=378
Score = 160 bits (404), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 72/106 (67%), Positives = 92/106 (86%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGF+VD ATLTRFF FHFILPFI++A+ ++HLLFLH+TGSNNP G++S++DKIPFHPY+T
Sbjct 167 GGFAVDNATLTRFFTFHFILPFIVLAMTMIHLLFLHQTGSNNPIGLNSNIDKIPFHPYFT 226
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KDI+G +++I LI LVL +P+LLGDPDN+ PA PL TP HI+PE
Sbjct 227 FKDIVGFIVMIFILISLVLISPNLLGDPDNFIPATPLVTPAHIQPE 272
> AtMi019
Length=393
Score = 130 bits (328), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 56/106 (52%), Positives = 81/106 (76%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVD ATL RFF+ H +LPFI++ +++HL LH+ GSNNP G+ S++DKI F+PY+
Sbjct 173 GGFSVDNATLNRFFSLHHLLPFILVGASLLHLAALHQYGSNNPLGVHSEMDKIAFYPYFY 232
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
+KD++G + + + + +AP++LG PDNY PANP++TPPHI PE
Sbjct 233 VKDLVGWVAFAIFFSIWIFYAPNVLGHPDNYIPANPMSTPPHIVPE 278
> At2g07727
Length=393
Score = 130 bits (328), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 56/106 (52%), Positives = 81/106 (76%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSVD ATL RFF+ H +LPFI++ +++HL LH+ GSNNP G+ S++DKI F+PY+
Sbjct 173 GGFSVDNATLNRFFSLHHLLPFILVGASLLHLAALHQYGSNNPLGVHSEMDKIAFYPYFY 232
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
+KD++G + + + + +AP++LG PDNY PANP++TPPHI PE
Sbjct 233 VKDLVGWVAFAIFFSIWIFYAPNVLGHPDNYIPANPMSTPPHIVPE 278
> HsMi013
Length=378
Score = 130 bits (328), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 83/106 (78%), Positives = 91/106 (85%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GG+SVD TLTRFF FHFILPFII A+A +HLLFLHETGSNNP GI+S DKI FHPYYT
Sbjct 166 GGYSVDSPTLTRFFTFHFILPFIIAALAALHLLFLHETGSNNPLGITSHSDKITFHPYYT 225
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
IKD LG LL +L+L+ L LF+PDLLGDPDNYT ANPLNTPPHIKPE
Sbjct 226 IKDALGLLLFLLSLMTLTLFSPDLLGDPDNYTLANPLNTPPHIKPE 271
> YMi014
Length=385
Score = 119 bits (297), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 55/106 (51%), Positives = 74/106 (69%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSV T+ RFFA H+++PFII A+ I+HL+ LH GS+NP GI+ ++D+IP H Y+
Sbjct 167 GGFSVSNPTIQRFFALHYLVPFIIAAMVIMHLMALHIHGSSNPLGITGNLDRIPMHSYFI 226
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KD++ L +L L L V ++P+ LG PDNY P NPL TP I PE
Sbjct 227 FKDLVTVFLFMLILALFVFYSPNTLGHPDNYIPGNPLVTPASIVPE 272
> SPMi004
Length=387
Score = 103 bits (257), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 73/106 (68%), Gaps = 0/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSV TL RFF+ H+++PF+I A++++HL+ LH GS+NP G+++++D+IP +PYY
Sbjct 167 GGFSVSNPTLNRFFSLHYLMPFVIAALSVMHLIALHTNGSSNPLGVTANMDRIPMNPYYL 226
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
IKD++ + ++ + + + P +PD PA+PL TP I PE
Sbjct 227 IKDLITIFIFLIGINYMAFYNPYGFMEPDCALPADPLKTPMSIVPE 272
> YMi015_1
Length=252
Score = 92.4 bits (228), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 42/86 (48%), Positives = 61/86 (70%), Gaps = 0/86 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GGFSV T+ RFFA H+++PFII A+ I+HL+ LH GS+NP GI+ ++D+IP H Y+
Sbjct 167 GGFSVSNPTIQRFFALHYLVPFIIAAMVIMHLMALHIHGSSNPLGITGNLDRIPMHSYFI 226
Query 61 IKDILGALLLILALILLVLFAPDLLG 86
KD++ L +L L L V ++P+ LG
Sbjct 227 FKDLVTVFLFMLILALFVFYSPNTLG 252
> CEMi012
Length=370
Score = 81.6 bits (200), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 45/106 (42%), Positives = 61/106 (57%), Gaps = 1/106 (0%)
Query 1 GGFSVDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYT 60
GF V ATL FF HF+LP+ I+ I + HL+FLH TGS + D DK+ F P Y
Sbjct 163 SGFGVTGATLKFFFVLHFLLPWAILVIVLGHLIFLHSTGSTSSLYCHGDYDKVCFSPEYL 222
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
KD ++ +L I+L L P LGD + + A+P+ +P HI PE
Sbjct 223 GKDAYNIVIWLL-FIVLSLIYPFNLGDAEMFIEADPMMSPVHIVPE 267
> Hs17442601
Length=234
Score = 62.4 bits (150), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 26/32 (81%), Positives = 30/32 (93%), Gaps = 0/32 (0%)
Query 75 ILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
+ L+LF+PDLLGDPDNYTPA+PLNTPPH KPE
Sbjct 203 LTLILFSPDLLGDPDNYTPASPLNTPPHTKPE 234
> Hs22041751
Length=210
Score = 60.5 bits (145), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 37/53 (69%), Positives = 43/53 (81%), Gaps = 0/53 (0%)
Query 5 VDKATLTRFFAFHFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHP 57
VDKATLTRFF FHF+L FII A+A VHL L ETGSNNP+G+SS+ +KI HP
Sbjct 158 VDKATLTRFFTFHFLLLFIITALAAVHLSCLRETGSNNPSGVSSNSNKITIHP 210
> Hs20537244
Length=290
Score = 53.9 bits (128), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 27/38 (71%), Positives = 29/38 (76%), Gaps = 0/38 (0%)
Query 69 LLILALILLVLFAPDLLGDPDNYTPANPLNTPPHIKPE 106
L I +L L VLF+PDLLGDP Y ANP NTPPHIKPE
Sbjct 240 LTIFSLHLAVLFSPDLLGDPGIYALANPFNTPPHIKPE 277
> Hs17458887
Length=212
Score = 48.5 bits (114), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 21/24 (87%), Positives = 22/24 (91%), Gaps = 0/24 (0%)
Query 83 DLLGDPDNYTPANPLNTPPHIKPE 106
DLL DPDNYT ANPLNTPPHIKP+
Sbjct 189 DLLSDPDNYTLANPLNTPPHIKPK 212
> AtCh052
Length=215
Score = 36.6 bits (83), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query 1 GGFSVDKATLTRFFAFH-FILPFIIIAIAIVHLLFLHETGSNNP 43
G SV ++TLTRF++ H F+LP + ++H L + + G + P
Sbjct 171 GSASVGQSTLTRFYSLHTFVLPLLTAVFMLMHFLMIRKQGISGP 214
> Hs11386155
Length=1173
Score = 30.4 bits (67), Expect = 0.76, Method: Composition-based stats.
Identities = 17/41 (41%), Positives = 26/41 (63%), Gaps = 1/41 (2%)
Query 61 IKDILGALLLILALILLVLFAPDLLGDPDNYTPANPLNTPP 101
+K+ILG + LA+I +LF +L D D+ A PL++PP
Sbjct 926 MKNILGHAVYQLAIIFTLLFVGELFFDIDSGRNA-PLHSPP 965
> 7298995
Length=555
Score = 30.4 bits (67), Expect = 0.91, Method: Composition-based stats.
Identities = 13/43 (30%), Positives = 26/43 (60%), Gaps = 6/43 (13%)
Query 37 ETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALILLVL 79
E +P G++ HPY+T++DIL + L++ +I+L++
Sbjct 474 EAQKMDPPGVTG------IHPYWTLRDILAFVTLLVVMIILLM 510
> At4g03470
Length=683
Score = 28.9 bits (63), Expect = 2.1, Method: Composition-based stats.
Identities = 20/64 (31%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Query 17 HFILPFIIIAIAIVHLLFLHETGSNNPTGISSDVDKIPFHPYYTIKDILGALLLILALIL 76
H+I + +A+V L F N+P + + P P KD + ALL++ ALI
Sbjct 454 HYIFRERLTLLALVQLHF-----QNDPRCAHTMIQTRPIMPQGGNKDYINALLVVAALIT 508
Query 77 LVLF 80
V F
Sbjct 509 TVTF 512
> SPCC737.06c
Length=287
Score = 27.3 bits (59), Expect = 7.5, Method: Compositional matrix adjust.
Identities = 14/52 (26%), Positives = 29/52 (55%), Gaps = 4/52 (7%)
Query 7 KATLTR-FFAFHFILPFIIIAIAIV---HLLFLHETGSNNPTGISSDVDKIP 54
+ TL++ F+ H + ++ +V H+ FL E+G+++ I +D+IP
Sbjct 100 EETLSQVFYNLHMLFGIDFVSTLVVSFPHITFLKESGNSSSNEIYDSIDEIP 151
Lambda K H
0.327 0.149 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1167556980
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40