bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0407_orf3
Length=213
Score E
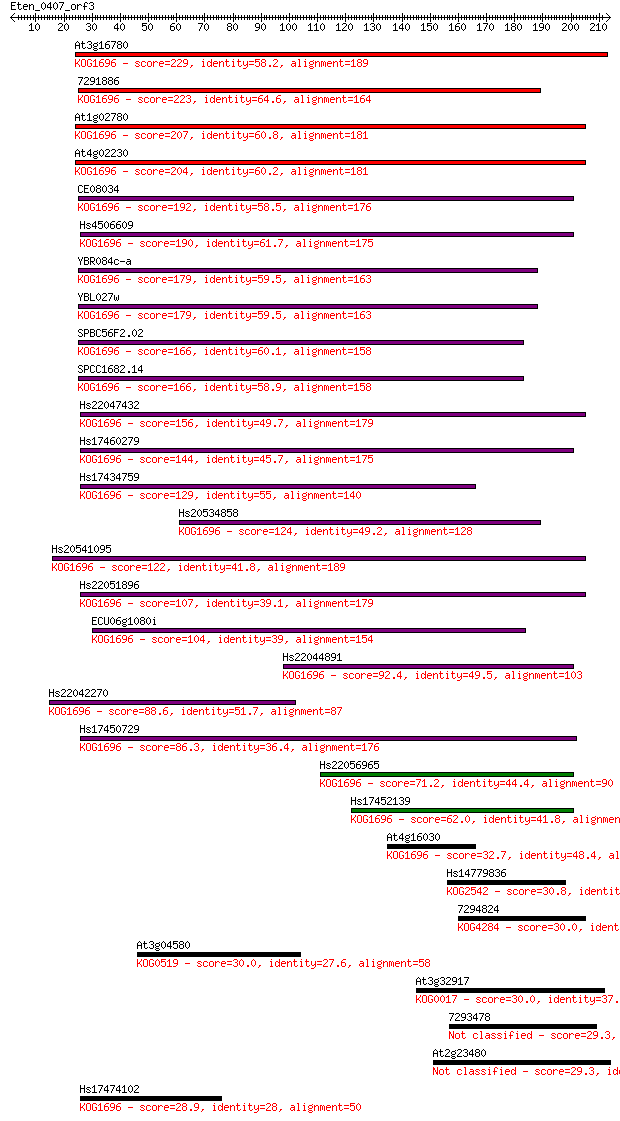
Sequences producing significant alignments: (Bits) Value
At3g16780 229 2e-60
7291886 223 2e-58
At1g02780 207 9e-54
At4g02230 204 1e-52
CE08034 192 6e-49
Hs4506609 190 2e-48
YBR084c-a 179 3e-45
YBL027w 179 3e-45
SPBC56F2.02 166 2e-41
SPCC1682.14 166 4e-41
Hs22047432 156 3e-38
Hs17460279 144 2e-34
Hs17434759 129 5e-30
Hs20534858 124 1e-28
Hs20541095 122 8e-28
Hs22051896 107 2e-23
ECU06g1080i 104 1e-22
Hs22044891 92.4 7e-19
Hs22042270 88.6 8e-18
Hs17450729 86.3 4e-17
Hs22056965 71.2 1e-12
Hs17452139 62.0 9e-10
At4g16030 32.7 0.49
Hs14779836 30.8 2.0
7294824 30.0 3.2
At3g04580 30.0 3.5
At3g32917 30.0 3.6
7293478 29.3 6.3
At2g23480 29.3 6.6
Hs17474102 28.9 8.0
> At3g16780
Length=209
Score = 229 bits (585), Expect = 2e-60, Method: Compositional matrix adjust.
Identities = 110/193 (56%), Positives = 146/193 (75%), Gaps = 4/193 (2%)
Query 24 MSLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKY 83
+SL++QKRLAAS++KCG+G+VWLDPNE+GDI+MANSR ++RKL++D IIRK +HS+
Sbjct 2 VSLKIQKRLAASVMKCGKGKVWLDPNESGDISMANSRQNIRKLVKDGFIIRKPTKIHSRS 61
Query 84 RFRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYH 143
R R +AKR+GRH G GKRKGTR+ARLP K+LWMRR RVLRR L KYR++KKID HMYH
Sbjct 62 RARALNEAKRKGRHSGYGKRKGTREARLPTKILWMRRMRVLRRFLSKYRESKKIDRHMYH 121
Query 144 KLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKK 203
+Y++ KGN FKNKRVL+E+IH K K +EK L +Q +A++ K A +E++ + E +
Sbjct 122 DMYMKVKGNVFKNKRVLMESIHKMKAEKAREKTLADQFEAKRIKNKASRERKFARREERL 181
Query 204 A----GVAAAKPA 212
A G PA
Sbjct 182 AQGPGGGETTTPA 194
> 7291886
Length=203
Score = 223 bits (568), Expect = 2e-58, Method: Compositional matrix adjust.
Identities = 106/164 (64%), Positives = 134/164 (81%), Gaps = 0/164 (0%)
Query 25 SLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYR 84
SL+LQKRLAAS+L+CG+ +VWLDPNE +IA NSR ++RKLI+D LII+K V VHS+YR
Sbjct 3 SLKLQKRLAASVLRCGKKKVWLDPNEINEIANTNSRQNIRKLIKDGLIIKKPVVVHSRYR 62
Query 85 FRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHK 144
R +A+R+GRH G GKRKGT +AR+P K+LWM+RQRVLRRLLKKYRD+KKID H+YH
Sbjct 63 VRKNTEARRKGRHCGFGKRKGTANARMPTKLLWMQRQRVLRRLLKKYRDSKKIDRHLYHD 122
Query 145 LYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKA 188
LY++CKGN FKNKRVL+E IH +K K + K L +Q +AR+QK
Sbjct 123 LYMKCKGNVFKNKRVLMEYIHKKKAEKQRSKMLADQAEARRQKV 166
> At1g02780
Length=214
Score = 207 bits (528), Expect = 9e-54, Method: Compositional matrix adjust.
Identities = 110/181 (60%), Positives = 142/181 (78%), Gaps = 0/181 (0%)
Query 24 MSLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKY 83
+SL+LQKRLAAS++KCG+G+VWLDPNE+ DI+MANSR ++RKL++D IIRK +HS+
Sbjct 2 VSLKLQKRLAASVMKCGKGKVWLDPNESSDISMANSRQNIRKLVKDGFIIRKPTKIHSRS 61
Query 84 RFRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYH 143
R R + AK +GRH G GKRKGTR+ARLP KVLWMRR RVLRRLLKKYR+ KKID HMYH
Sbjct 62 RARKMKIAKMKGRHSGYGKRKGTREARLPTKVLWMRRMRVLRRLLKKYRETKKIDKHMYH 121
Query 144 KLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKK 203
+Y+R KGN FKNKRVL+E+IH K K +EK L +Q +A++ K A +E++ + E +
Sbjct 122 DMYMRVKGNVFKNKRVLMESIHKSKAEKAREKTLSDQFEAKRAKNKASRERKHARREERL 181
Query 204 A 204
A
Sbjct 182 A 182
> At4g02230
Length=208
Score = 204 bits (519), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 109/181 (60%), Positives = 140/181 (77%), Gaps = 0/181 (0%)
Query 24 MSLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKY 83
+SL+LQKRLA+S+LKCG+ +VWLDPNE DI+MANSR ++RKL++D IIRK +HS+
Sbjct 2 VSLKLQKRLASSVLKCGKRKVWLDPNEGSDISMANSRQNIRKLVKDGFIIRKPTKIHSRS 61
Query 84 RFRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYH 143
R R AKR+GRH G GKRKGTR+ARLP KVLWMRR RVLRRLLKKYR+ KKID HMYH
Sbjct 62 RARQLNIAKRKGRHSGYGKRKGTREARLPTKVLWMRRMRVLRRLLKKYRETKKIDRHMYH 121
Query 144 KLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKK 203
+Y++ KGN FKNKRVL+E+IH K K +EK L +Q +A++ K A +E++ + E +
Sbjct 122 DMYMKVKGNVFKNKRVLMESIHKSKAEKAREKTLSDQFEAKRAKNKASRERKHARREERL 181
Query 204 A 204
A
Sbjct 182 A 182
> CE08034
Length=198
Score = 192 bits (487), Expect = 6e-49, Method: Compositional matrix adjust.
Identities = 103/176 (58%), Positives = 135/176 (76%), Gaps = 3/176 (1%)
Query 25 SLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYR 84
+LRLQKRLA+++LKCG+ RVWLDPNE +I+ ANSR S+R+L+ D LIIRK V VHS++R
Sbjct 3 NLRLQKRLASAVLKCGKHRVWLDPNEVSEISGANSRQSIRRLVNDGLIIRKPVTVHSRFR 62
Query 85 FRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHK 144
R Y++A+R+GRH G GKR+GT +AR+P K LW+RR RVLR LL++YRDAKK+D H+YH+
Sbjct 63 AREYEEARRKGRHTGYGKRRGTANARMPEKTLWIRRMRVLRNLLRRYRDAKKLDKHLYHE 122
Query 145 LYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 200
LY+R KGN FKNK+ LIE I +K + K L +Q AR+ K KE RK +EE
Sbjct 123 LYLRAKGNNFKNKKNLIEYIFKKKTENKRAKQLADQAQARRDK---NKESRKRREE 175
> Hs4506609
Length=196
Score = 190 bits (483), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 108/175 (61%), Positives = 136/175 (77%), Gaps = 3/175 (1%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRF 85
LRLQKRLA+S+L+CG+ +VWLDPNE +IA ANSR +RKLI+D LIIRK V VHS+ R
Sbjct 4 LRLQKRLASSVLRCGKKKVWLDPNETNEIANANSRQQIRKLIKDGLIIRKPVTVHSRARC 63
Query 86 RLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKL 145
R A+R+GRHMGIGKRKGT +AR+P KV WMRR R+LRRLL++YR++KKID HMYH L
Sbjct 64 RKNTLARRKGRHMGIGKRKGTANARMPEKVTWMRRMRILRRLLRRYRESKKIDRHMYHSL 123
Query 146 YVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 200
Y++ KGN FKNKR+L+E IH K K ++K L +Q +AR+ K KE RK +EE
Sbjct 124 YLKVKGNVFKNKRILMEHIHKLKADKARKKLLADQAEARRSKT---KEARKRREE 175
> YBR084c-a
Length=189
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 97/163 (59%), Positives = 125/163 (76%), Gaps = 0/163 (0%)
Query 25 SLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYR 84
+LR QKRLAAS++ G+ +VWLDPNE +IA ANSR ++RKL+++ I++KAV VHSK R
Sbjct 3 NLRTQKRLAASVVGVGKRKVWLDPNETSEIAQANSRNAIRKLVKNGTIVKKAVTVHSKSR 62
Query 85 FRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHK 144
R + Q+KR+GRH G GKRKGTR+ARLP++V+W+RR RVLRRLL KYRDA KID H+YH
Sbjct 63 TRAHAQSKREGRHSGYGKRKGTREARLPSQVVWIRRLRVLRRLLAKYRDAGKIDKHLYHV 122
Query 145 LYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQK 187
LY KGN FK+KR L+E I K +EKAL E+ +AR+ K
Sbjct 123 LYKESKGNAFKHKRALVEHIIQAKADAQREKALNEEAEARRLK 165
> YBL027w
Length=189
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 97/163 (59%), Positives = 125/163 (76%), Gaps = 0/163 (0%)
Query 25 SLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYR 84
+LR QKRLAAS++ G+ +VWLDPNE +IA ANSR ++RKL+++ I++KAV VHSK R
Sbjct 3 NLRTQKRLAASVVGVGKRKVWLDPNETSEIAQANSRNAIRKLVKNGTIVKKAVTVHSKSR 62
Query 85 FRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHK 144
R + Q+KR+GRH G GKRKGTR+ARLP++V+W+RR RVLRRLL KYRDA KID H+YH
Sbjct 63 TRAHAQSKREGRHSGYGKRKGTREARLPSQVVWIRRLRVLRRLLAKYRDAGKIDKHLYHV 122
Query 145 LYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQK 187
LY KGN FK+KR L+E I K +EKAL E+ +AR+ K
Sbjct 123 LYKESKGNAFKHKRALVEHIIQAKADAQREKALNEEAEARRLK 165
> SPBC56F2.02
Length=193
Score = 166 bits (421), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 95/158 (60%), Positives = 120/158 (75%), Gaps = 0/158 (0%)
Query 25 SLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYR 84
+LR QKRLAAS+LKCG+ +VW+DPNE +I+ ANSR +VRKLI+D L+IRK +HS++R
Sbjct 3 NLRTQKRLAASVLKCGKRKVWMDPNEISEISNANSRQNVRKLIKDGLVIRKPNLMHSRFR 62
Query 85 FRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHK 144
R AKR GRH G GKRKGT +AR+P+ V+WMRRQRVLRRLL+KYR++ KID H+YH
Sbjct 63 IRKTHAAKRLGRHTGYGKRKGTAEARMPSAVVWMRRQRVLRRLLRKYRESGKIDKHLYHT 122
Query 145 LYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVD 182
LY+ KGN FK+KR LIE I K + K +QEQ D
Sbjct 123 LYLEAKGNTFKHKRALIEHIQRAKAEANRTKLIQEQQD 160
> SPCC1682.14
Length=193
Score = 166 bits (419), Expect = 4e-41, Method: Compositional matrix adjust.
Identities = 93/158 (58%), Positives = 120/158 (75%), Gaps = 0/158 (0%)
Query 25 SLRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYR 84
+LR QKRLAAS+LKCG+ +VW+DPNE +I+ ANSR ++RKL++D L+IRK +HS++R
Sbjct 3 NLRTQKRLAASVLKCGKRKVWMDPNEISEISNANSRQNIRKLVKDGLVIRKPNLMHSRFR 62
Query 85 FRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHK 144
R AKR GRH G GKRKGT +AR+P+ V+WMRRQRVLRRLL+KYR++ KID H+YH
Sbjct 63 IRKTHAAKRLGRHTGYGKRKGTAEARMPSTVVWMRRQRVLRRLLRKYRESGKIDKHLYHT 122
Query 145 LYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVD 182
LY+ KGN FK+KR LIE I K + K +QEQ D
Sbjct 123 LYLEAKGNTFKHKRALIEHIQRAKAEANRTKLIQEQQD 160
> Hs22047432
Length=521
Score = 156 bits (394), Expect = 3e-38, Method: Compositional matrix adjust.
Identities = 89/179 (49%), Positives = 117/179 (65%), Gaps = 16/179 (8%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRF 85
L LQKR AAS+L+ G+ +VWLDPNEA +IA ANSR +RKLI+D LII K V VHS+ +
Sbjct 4 LGLQKRPAASVLRYGKKKVWLDPNEANEIASANSRQQIRKLIKDGLIICKPVTVHSQAQC 63
Query 86 RLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKL 145
R A R+GRHMG GKRKGT +A +P KV +YRD+KKI+ HMYH L
Sbjct 64 RKNTLAHRKGRHMGTGKRKGTANAPMPEKV-------------TRYRDSKKINHHMYHSL 110
Query 146 YVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 204
Y++ K + FK+K++L+E IH K K +K L +Q +A + K KE RK +EE +A
Sbjct 111 YLKVKESVFKDKQILMEHIHKLKADKAGKKLLADQAEACRPKT---KEARKQREECLQA 166
> Hs17460279
Length=170
Score = 144 bits (362), Expect = 2e-34, Method: Compositional matrix adjust.
Identities = 80/175 (45%), Positives = 106/175 (60%), Gaps = 29/175 (16%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRF 85
LRLQKRLA+S+L+CG+ +VWLDPN+ +IA ANS + KLI+D LIIRK V
Sbjct 4 LRLQKRLASSVLRCGKKKVWLDPNKTNEIANANSHQQIWKLIKDGLIIRKPVT------- 56
Query 86 RLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKL 145
GT +AR+P KV WM+R R+L LL++Y ++KKID HMYH L
Sbjct 57 -------------------GTANARMPEKVTWMKRMRILHLLLRRYCESKKIDRHMYHSL 97
Query 146 YVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 200
Y++ +GN F NK +L+E H K K +K L +Q +AR+ K KE RK EE
Sbjct 98 YLKVQGNVFTNKPILMEHSHKLKADKAHKKLLADQAEARRPKT---KEARKRSEE 149
> Hs17434759
Length=138
Score = 129 bits (323), Expect = 5e-30, Method: Compositional matrix adjust.
Identities = 77/140 (55%), Positives = 97/140 (69%), Gaps = 8/140 (5%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRF 85
LRLQKRLA+SIL CG+ +VWLDP E +IA NSR +RKLIRD LII K V VHS
Sbjct 4 LRLQKRLASSILCCGKKKVWLDPGETSEIANVNSRQQIRKLIRDGLIILKPVPVHS---- 59
Query 86 RLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKL 145
QA+ + K+KGT +AR+P K+ WMRR R+LR LL++Y ++KKID +MYH L
Sbjct 60 ----QAQCWKNTLAGRKQKGTANARMPEKITWMRRMRILRLLLRRYCESKKIDCYMYHSL 115
Query 146 YVRCKGNQFKNKRVLIEAIH 165
Y++ KGN FKNK +L E H
Sbjct 116 YLKVKGNVFKNKWILREHSH 135
> Hs20534858
Length=376
Score = 124 bits (311), Expect = 1e-28, Method: Compositional matrix adjust.
Identities = 63/128 (49%), Positives = 89/128 (69%), Gaps = 2/128 (1%)
Query 61 FSVRKLIRDNLIIRKAVAVHSKYRFRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRR 120
++KLI+D LII K V VHS+ + A R+GRHMGIGK+K T +A++P KV WMR+
Sbjct 251 LEIQKLIKDGLIIHKPVIVHSQAQCWKNTLACRKGRHMGIGKQKRTANAQIPEKVTWMRK 310
Query 121 QRVLRRLLKKYRDAKKIDSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQ 180
+L RLL++YR++KKID HMYH LY++ KGN FKNK +++E I+ K K +K L +Q
Sbjct 311 --ILHRLLRRYRESKKIDGHMYHSLYLKVKGNVFKNKWIVMEHINKLKADKAHKKLLADQ 368
Query 181 VDARKQKA 188
+A + K
Sbjct 369 AEAPRSKT 376
> Hs20541095
Length=240
Score = 122 bits (305), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 79/190 (41%), Positives = 110/190 (57%), Gaps = 35/190 (18%)
Query 16 PPSSAQGKMS-LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIR 74
P ++A MS LRLQKRLA+S+L G+ ++ RKLI+D LII
Sbjct 68 PFTAAATAMSMLRLQKRLASSVLHWGKKKI-------------------RKLIKDGLIIW 108
Query 75 KAVAVHSKYRFRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDA 134
K V VHS+ +F A ++G+HM IG+ V W+RR R+L RLL++Y ++
Sbjct 109 KPVTVHSQAQFWKNTLAHQKGKHMCIGQ------------VTWIRRTRILCRLLRRYHES 156
Query 135 KKIDSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEK 194
KKID HMYH LY++ KGN FKNK++L+E I K K +K L +Q +AR+ K K+
Sbjct 157 KKIDCHMYHSLYLQMKGNVFKNKQILMEYIRKLKAGKACKKLLADQAEARRSKT---KDA 213
Query 195 RKLKEENKKA 204
RK EE +A
Sbjct 214 RKHSEERLQA 223
> Hs22051896
Length=145
Score = 107 bits (267), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 70/179 (39%), Positives = 92/179 (51%), Gaps = 47/179 (26%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRF 85
L LQKRLA+S+L+CG+ +VWLDP E +IA ANS +RKLI+D LII K H Y
Sbjct 4 LSLQKRLASSVLRCGKKKVWLDPKETSEIANANSHQWIRKLIKDGLIIHKPKIDHHMYHR 63
Query 86 RLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKL 145
R Y ++K+ RH MYH L
Sbjct 64 R-YHESKKVDRH-------------------------------------------MYHSL 79
Query 146 YVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 204
Y++ KGN FKNKR+L+E IHN K K +K L +Q + R+ K KE RK +EE+ +A
Sbjct 80 YLKVKGNVFKNKRILMEHIHNLKADKACKKLLADQAETRRSKI---KEARKHREEHLQA 135
> ECU06g1080i
Length=171
Score = 104 bits (260), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 60/154 (38%), Positives = 95/154 (61%), Gaps = 0/154 (0%)
Query 30 KRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRFRLYQ 89
+RLA+ IL CG+ ++WLD NE + A++R +++LI+DN+II K S+ R R
Sbjct 8 RRLASDILNCGKKKLWLDNNEIERLNGASTREQIKQLIKDNVIIMKQDKHTSRGRCRKRL 67
Query 90 QAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKLYVRC 149
+AK++GRHMG GKR GT +AR+P K +WM++ R +R +LK+ R +I + Y +
Sbjct 68 EAKKKGRHMGTGKRFGTANARMPQKKIWMKKIRAMRAMLKEMRANGEITKEDFRVFYKQA 127
Query 150 KGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDA 183
KG+ FK++ + + I K + + K L EQ A
Sbjct 128 KGHLFKHRFHMKDCIIKRKADEQRAKELAEQAKA 161
> Hs22044891
Length=120
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 51/103 (49%), Positives = 67/103 (65%), Gaps = 3/103 (2%)
Query 98 MGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKLYVRCKGNQFKNK 157
MGI KRKGT +A++P V WMRR R+L LL++Y ++KKID H YH LY++ KGN FKNK
Sbjct 1 MGISKRKGTANAQMPGNVTWMRRMRILCWLLRRYCESKKIDHHTYHSLYLKVKGNVFKNK 60
Query 158 RVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEE 200
+L+E I K K +K +Q AR+ K KE RK E+
Sbjct 61 WILMEHILKLKADKAHKKLQADQAKARRSKT---KEARKHHED 100
> Hs22042270
Length=383
Score = 88.6 bits (218), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 45/88 (51%), Positives = 61/88 (69%), Gaps = 1/88 (1%)
Query 15 APPSSAQGKMS-LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLII 73
+P ++ MS LRLQKRLA+ +L CG+ +VWLDPNE +I+ ANS + KLI+D L+I
Sbjct 236 SPFTAMVTAMSMLRLQKRLASRVLPCGKKKVWLDPNETNEISNANSCQQIWKLIKDGLMI 295
Query 74 RKAVAVHSKYRFRLYQQAKRQGRHMGIG 101
RK + VHS+ R + A R+GRHMGI
Sbjct 296 RKPMTVHSQARCQENTLAYRKGRHMGIA 323
> Hs17450729
Length=147
Score = 86.3 bits (212), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 64/178 (35%), Positives = 87/178 (48%), Gaps = 56/178 (31%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIR--KAVAVHSKY 83
L QKRL +S+L CG ++ RKLI+D LI+R K V VHS+
Sbjct 4 LTPQKRLISSVLHCGEKKI-------------------RKLIKDGLILRHRKPVTVHSRA 44
Query 84 RFRLYQQAKRQGRHMGIGKRKGTRDARLPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYH 143
+ A+R+GRH+GI ++KKID HMYH
Sbjct 45 QCWKSTLARRKGRHLGI--------------------------------ESKKIDRHMYH 72
Query 144 KLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEEN 201
LY++ KGN FK+KR+L E H K K ++K L +Q +AR K KE RKL+EE+
Sbjct 73 SLYLKLKGNVFKHKRILTEHSHKLKADKARKKPLADQAEARGSKT---KEARKLREEH 127
> Hs22056965
Length=108
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 40/90 (44%), Positives = 56/90 (62%), Gaps = 3/90 (3%)
Query 111 LPAKVLWMRRQRVLRRLLKKYRDAKKIDSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNL 170
+ K WMRR R+L RLL++Y ++KK D H+Y LY++ KGN FKNK++ E IH K
Sbjct 1 MSQKDTWMRRMRILCRLLRRYLESKKTDCHVYPSLYLKVKGNVFKNKQIFTEHIHKLKTD 60
Query 171 KVKEKALQEQVDARKQKAAAQKEKRKLKEE 200
+ +K L +Q AR+ K KE+ K EE
Sbjct 61 EAHKKLLADQAVARRSKT---KEEHKHHEE 87
> Hs17452139
Length=118
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 33/79 (41%), Positives = 48/79 (60%), Gaps = 3/79 (3%)
Query 122 RVLRRLLKKYRDAKKIDSHMYHKLYVRCKGNQFKNKRVLIEAIHNEKNLKVKEKALQEQV 181
R+L L +Y ++KK D HMYH LY++ +GN F NK++L+E H K K +K L +Q
Sbjct 2 RILHWLFGRYHESKKTDHHMYHSLYLKVQGNMFTNKQILMEHNHKLKADKAHKKFLADQA 61
Query 182 DARKQKAAAQKEKRKLKEE 200
+AR K + KL+EE
Sbjct 62 EARSSKT---NKASKLREE 77
> At4g16030
Length=101
Score = 32.7 bits (73), Expect = 0.49, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 24/32 (75%), Gaps = 1/32 (3%)
Query 135 KKIDSHMY-HKLYVRCKGNQFKNKRVLIEAIH 165
KKID +Y H ++++ KG +KNK VL+E++H
Sbjct 29 KKIDKLVYYHDMFMKVKGKVYKNKCVLMESMH 60
> Hs14779836
Length=509
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/42 (42%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 156 NKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKL 197
N R L EA+ E L++ E L E+ DA Q+ QK +RKL
Sbjct 228 NLRKLAEALQPELPLELGEAILAEEFDAEFQRHYLQKMRRKL 269
> 7294824
Length=1028
Score = 30.0 bits (66), Expect = 3.2, Method: Composition-based stats.
Identities = 17/45 (37%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 160 LIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKA 204
L E+ HN KNL+VKEK + Q ++ + A K E KK+
Sbjct 488 LPESSHNNKNLRVKEKEIVRQGKLKETNSKAGGSKTNKTERRKKS 532
> At3g04580
Length=766
Score = 30.0 bits (66), Expect = 3.5, Method: Composition-based stats.
Identities = 16/58 (27%), Positives = 28/58 (48%), Gaps = 0/58 (0%)
Query 46 LDPNEAGDIAMANSRFSVRKLIRDNLIIRKAVAVHSKYRFRLYQQAKRQGRHMGIGKR 103
+ P + G A+ RF + LIR+ + K ++V+ Y F + Q + +G KR
Sbjct 426 ISPKDNGKSALEVKRFQLHSLIREAACVAKCLSVYKGYGFEMDVQTRLPNLVVGDEKR 483
> At3g32917
Length=757
Score = 30.0 bits (66), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 25/77 (32%), Positives = 39/77 (50%), Gaps = 11/77 (14%)
Query 145 LYVRCKGNQFKNKRVLIE-AIHNEKNLK---------VKEKALQEQVDARKQKAAAQKEK 194
L VRC+ +Q+ K L+E A+ E++L+ V+ K Q+QV K AQ +K
Sbjct 265 LRVRCRVSQYATKAALVETAVKVEEDLQRQVVAVSRAVQPKKTQQQVAPSKGNKPAQGQK 324
Query 195 RKLKEENKKAGVAAAKP 211
RK + +AG +P
Sbjct 325 RKW-DHPSRAGQGHLRP 340
> 7293478
Length=1004
Score = 29.3 bits (64), Expect = 6.3, Method: Composition-based stats.
Identities = 17/52 (32%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 157 KRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKAGVAA 208
K +L I E+ +K++EK QE V+ + K Q K+K K + A A
Sbjct 762 KEILTSKIDEEEMIKLQEKIKQEIVNTKFFKGEQQPRKKKRKNQRAAADAPA 813
> At2g23480
Length=705
Score = 29.3 bits (64), Expect = 6.6, Method: Composition-based stats.
Identities = 21/64 (32%), Positives = 33/64 (51%), Gaps = 1/64 (1%)
Query 151 GNQFKNKRVLIEAIHNEKNLKVKEKALQEQVDARKQKAAAQKEKRKLKEENKKAGV-AAA 209
G Q K K + A + +K + ++E+V A+K K A +K K++ KK V A A
Sbjct 8 GGQGKRKEIEAPAPAKTEKVKAPAEKVKEKVPAKKAKVQAPAKKEKVQAPAKKEKVQAPA 67
Query 210 KPAQ 213
K A+
Sbjct 68 KKAK 71
> Hs17474102
Length=105
Score = 28.9 bits (63), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 14/50 (28%), Positives = 28/50 (56%), Gaps = 0/50 (0%)
Query 26 LRLQKRLAASILKCGRGRVWLDPNEAGDIAMANSRFSVRKLIRDNLIIRK 75
LRLQ++ ++++ CG+ + LD + + + R V+K + I+RK
Sbjct 4 LRLQRKFSSNVFCCGKKKKTLDHRKGRHLGIEGERDCVQKQAYSHGIVRK 53
Lambda K H
0.319 0.131 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3897491854
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40