bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
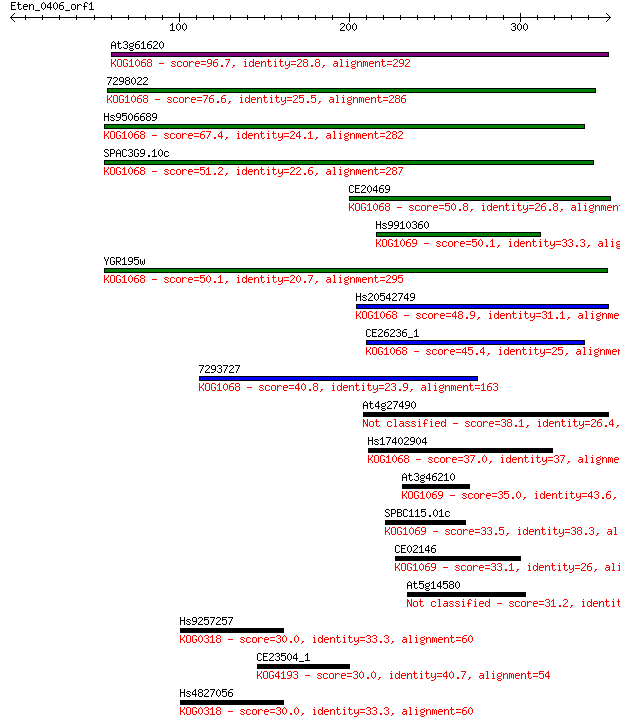
Query= Eten_0406_orf1
Length=352
Score E
Sequences producing significant alignments: (Bits) Value
At3g61620 96.7 8e-20
7298022 76.6 8e-14
Hs9506689 67.4 5e-11
SPAC3G9.10c 51.2 3e-06
CE20469 50.8 4e-06
Hs9910360 50.1 7e-06
YGR195w 50.1 8e-06
Hs20542749 48.9 1e-05
CE26236_1 45.4 2e-04
7293727 40.8 0.004
At4g27490 38.1 0.031
Hs17402904 37.0 0.068
At3g46210 35.0 0.22
SPBC115.01c 33.5 0.76
CE02146 33.1 0.91
At5g14580 31.2 3.4
Hs9257257 30.0 8.0
CE23504_1 30.0 8.5
Hs4827056 30.0 8.9
> At3g61620
Length=241
Score = 96.7 bits (239), Expect = 8e-20, Method: Compositional matrix adjust.
Identities = 84/294 (28%), Positives = 126/294 (42%), Gaps = 72/294 (24%)
Query 60 EAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSLASCD 119
E V+P G R DGR E RQ+ AEV G ++ D
Sbjct 2 EYVNPEGLRLDGRRFNEMRQI----------------VAEV------------GVVSKAD 33
Query 120 GRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLAR 179
G AV +G T V+A VYGP+ N S N +V
Sbjct 34 GSAVFEMGNTKVIAAVYGPREIQNKSQ---QKKNDHAV---------------------- 68
Query 180 GELAVPVSVVC--SVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITIS 237
V+C S+ + R R ++ + E++ +R E IL L ++I I
Sbjct 69 --------VLCEYSMAQFSTGDRRRQKFDRRSTELSLVIRQTMEACILTELMPHSQIDIF 120
Query 238 ILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTS 297
+ VL DG SA + AA+LALADAG+ MRDL SC+ L PLL ++
Sbjct 121 LQVLQADGGTRSACINAATLALADAGIPMRDLAVSCSAGYL-------NSTPLLDLNYVE 173
Query 298 DETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECMK 351
D +GG +T+G+ + + + LQ+ ++ +T+E +F + GC A AE ++
Sbjct 174 DS--AGGADVTVGILPKLDKVSLLQMDAKLPMETFETVFALASEGCKAIAERIR 225
> 7298022
Length=246
Score = 76.6 bits (187), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 73/290 (25%), Positives = 108/290 (37%), Gaps = 79/290 (27%)
Query 58 KAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSLAS 117
K + +S G R DGR +E R+++C+ V E PDG A
Sbjct 4 KYDLLSEQGLRLDGRRPHELRRIKCKLGV-----------FEQPDGSA------------ 40
Query 118 CDGRAVVSLGCTXVMALVYGPQPSSNMSTLST----SSSNTWSVSCTDNTSAAMQMMHEG 173
+ G T V+A VYGP + T S S T+S + N
Sbjct 41 -----YMEQGNTKVLAAVYGPHQAKGNQTESVINCQYSQATFSTAERKN----------- 84
Query 174 KQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTR 233
R R + E ++ A I LY +++
Sbjct 85 ----------------------------RPRGDRKSLEFKLYLQQALSAAIKSELYPRSQ 116
Query 234 ITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLV 293
I I + VL DDG+ + +L AA+LAL DAG+ + + + +CT L P L
Sbjct 117 IDIYVEVLQDDGANYAVALNAATLALIDAGICLNEFIVACTASLSKSNIP--------LT 168
Query 294 DPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGC 343
D + E SGGP LT+ I L+++ R E + E + GC
Sbjct 169 DISQFEEVSGGPKLTVAALPTAEKIAFLEMSERFHIDHLETVIETAMAGC 218
> Hs9506689
Length=245
Score = 67.4 bits (163), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 68/285 (23%), Positives = 109/285 (38%), Gaps = 73/285 (25%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M E +S G+R DGR E R+++ R V
Sbjct 1 MAGLELLSDQGYRVDGRRAGELRKIQARMGV----------------------------F 32
Query 116 ASCDGRAVVSLGCTXVMALVYGPQP---SSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHE 172
A DG A + G T +A+VYGP S + + N S T +T + H
Sbjct 33 AQADGSAYIEQGNTKALAVVYGPHEIRGSRARALPDRALVNCQYSSATFSTGERKRRPHG 92
Query 173 GKQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQT 232
++ C +GL +R E IL +L+ ++
Sbjct 93 DRKS-------------CEMGL--------------------QLRQTFEAAILTQLHPRS 119
Query 233 RITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLL 292
+I I + VL DG +A + AA+LA+ DAG+ MRD + +C+ + + L
Sbjct 120 QIDIYVQVLQADGGTYAACVNAATLAVLDAGIPMRDFVCACSAGFV---------DGTAL 170
Query 293 VDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFE 337
D + E +GGP L L + + I L++ R+ E++ E
Sbjct 171 ADLSHVEEAAGGPQLALALLPASGQIALLEMDARLHEDHLERVLE 215
> SPAC3G9.10c
Length=242
Score = 51.2 bits (121), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 65/288 (22%), Positives = 112/288 (38%), Gaps = 71/288 (24%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M E +S G R DGR E R +CR +
Sbjct 1 MSHFEILSLEGLRNDGRRWDEMRNFQCRIGIE---------------------------- 32
Query 116 ASCDGRAVVSLGCTXVMALVYGP-QPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGK 174
S +G A + LG T V+ +V GP +P + + K
Sbjct 33 PSENGSAFIELGNTKVLCIVDGPSEP-----------------------------VIKSK 63
Query 175 QGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRI 234
R + V +++ S +D V+ R + A++ E II LY +++I
Sbjct 64 ARADRTFVNVEINI-ASFSTID--VKKRFKSDRRIQLQCLALQNTFEEIIQTELYPRSQI 120
Query 235 TISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVD 294
++ + VL DDG+++++ + A +LAL DAG+ ++D + T ++ E +L+D
Sbjct 121 SVYLHVLQDDGAVMASCINATTLALIDAGIPVKDFVCCSTAGIV---------ESDMLLD 171
Query 295 PTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRG 342
S E S LT+ V +V +QL + E + + + G
Sbjct 172 LNSLE-ESALSWLTVAVLGNIKKVVYMQLETSMHLDYLESVMNMAIAG 218
> CE20469
Length=240
Score = 50.8 bits (120), Expect = 4e-06, Method: Compositional matrix adjust.
Identities = 41/153 (26%), Positives = 73/153 (47%), Gaps = 9/153 (5%)
Query 200 RTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLAL 259
+ R+R + EI+ + A E +IL + ++++ I V+ DGS L+A + A SLAL
Sbjct 82 KNRTRGDRKSTEISRLLEKAFESVILTEAFPRSQLDIFCEVIQGDGSNLAACVNATSLAL 141
Query 260 ADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIV 319
ADAG+ M+ + + T ++ + P +VD TS E P +TL + ++
Sbjct 142 ADAGIPMKGIASAATCGVVDGK-P--------IVDLTSREETDLLPRVTLATICGRDEVI 192
Query 320 ALQLTGRVDRQTYEKMFEVCLRGCLATAECMKV 352
++L R+ + + C EC+ V
Sbjct 193 LVELQNRLHIDHLSTVMDAAKATCADVYECLAV 225
> Hs9910360
Length=235
Score = 50.1 bits (118), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 50/96 (52%), Gaps = 8/96 (8%)
Query 216 VRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTV 275
+R E ++L L+ +T IT+ + V++D GS+L+ L AA +AL DAGV MR L
Sbjct 98 IRNTCEAVVLGTLHPRTSITVVLQVVSDAGSLLACCLNAACMALVDAGVPMRALFCGVAC 157
Query 276 LLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGV 311
L + L++DPTS + + LT +
Sbjct 158 AL--------DSDGTLVLDPTSKQEKEARAVLTFAL 185
> YGR195w
Length=246
Score = 50.1 bits (118), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 61/297 (20%), Positives = 106/297 (35%), Gaps = 71/297 (23%)
Query 56 MFKAEAVSPCGFRADGRLVYECRQLRCRTSVPSAAASDLTTSAEVPDGPAIATAGASGSL 115
M + E SP G R DGR E R+ + AA
Sbjct 1 MSRLEIYSPEGLRLDGRRWNELRRFESSINTHPHAA------------------------ 36
Query 116 ASCDGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMHEGKQ 175
DG + + G ++ LV GP+ S + TS +
Sbjct 37 ---DGSSYMEQGNNKIITLVKGPKEPRLKSQMDTSKA----------------------- 70
Query 176 GLARGELAVPVSVVCSVGLVDSCVRTRSRYTPD--AAEIAAAVRTAAEGIILRRLYTQTR 233
++V ++ R++S + + EI ++ E ++ +Y +T
Sbjct 71 ---------LLNVSVNITKFSKFERSKSSHKNERRVLEIQTSLVRMFEKNVMLNIYPRTV 121
Query 234 ITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLV 293
I I I VL DG I+ + + +LAL DAG++M D + +V L PLL
Sbjct 122 IDIEIHVLEQDGGIMGSLINGITLALIDAGISMFDYISGISVGLYDTT-------PLLDT 174
Query 294 DPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECM 350
+ + S +TLGV ++ + L + ++ E + + + G + M
Sbjct 175 NSLEENAMS---TVTLGVVGKSEKLSLLLVEDKIPLDRLENVLAIGIAGAHRVRDLM 228
> Hs20542749
Length=206
Score = 48.9 bits (115), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 46/161 (28%), Positives = 67/161 (41%), Gaps = 25/161 (15%)
Query 204 RYTP----DAAEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLAL 259
R+ P + E A ++ A E + Y ++ S L+L D GS L+ L A LAL
Sbjct 52 RHAPTGGGEEPEPAMVLQEALEPAMCLDRYPHVQLEASELLLEDGGSALATELNTAKLAL 111
Query 260 ADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIV 319
A AG+ M DL+ SC + L P P L+DP E P +TL + +
Sbjct 112 AYAGMEMNDLVVSCGLGLTPGSVPT------WLLDPMRLEEEHTAPGVTLAL------LP 159
Query 320 ALQLTGRVDRQTYE---------KMFEVCLRGCLATAECMK 351
L GR D + + + L+ CL A C +
Sbjct 160 MLNQGGRPDHELGGGCRPGPRGLRALDPMLQQCLVWAACHR 200
> CE26236_1
Length=252
Score = 45.4 bits (106), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 56/128 (43%), Gaps = 8/128 (6%)
Query 210 AEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDL 269
AE A + +A +I Y I I I VL+DDG +LS +L A +LA++ +G+ L
Sbjct 97 AEYRAQLASAVSAVIFASKYPGKVIEIEITVLSDDGGVLSTALSAVTLAISHSGIENMGL 156
Query 270 LPSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDR 329
+ S V + + L DP++ E+ +T + + GR+
Sbjct 157 MASVHVAM--------NSDGECLTDPSTSESEGAIGGVTFAFVPNLGQTTCVDIYGRIPL 208
Query 330 QTYEKMFE 337
++ + E
Sbjct 209 KSTSSLLE 216
> 7293727
Length=326
Score = 40.8 bits (94), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 39/163 (23%), Positives = 70/163 (42%), Gaps = 34/163 (20%)
Query 112 SGSLASCDGRAVVSLGCTXVMALVYGPQPSSNMSTLSTSSSNTWSVSCTDNTSAAMQMMH 171
+G L + G A + G T V+A+V P + S N ++C N +A
Sbjct 54 AGVLTTVRGSAYMEYGNTKVLAIV---APPKELIRASARRMNMGVLNCYVNFAA------ 104
Query 172 EGKQGLARGELAVPVSVVCSVGLVDSCVRTRSRYTPDAAEIAAAVRTAAEGIILRRLYTQ 231
S G +DS V R R+ +++ + A E ++ R +
Sbjct 105 ------------------FSTGDLDS-VPERERH------LSSMLTKAMEPVVCRTEFLN 139
Query 232 TRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCT 274
++ I +L+L DDG +LS ++ +AL + G++ DL+ + T
Sbjct 140 FQLDIRVLILDDDGCLLSTAINCCGVALVECGISTYDLITAST 182
> At4g27490
Length=244
Score = 38.1 bits (87), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 38/181 (20%), Positives = 64/181 (35%), Gaps = 46/181 (25%)
Query 208 DAAEIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSI-------------------- 247
D E ++ + A EG+I+ + +T + + LVL G
Sbjct 61 DHKEYSSMLHKALEGVIMMETFPKTTVDVFALVLESGGKCDCLLVRLDIRGVDYGSNLLD 120
Query 248 -----------------LSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPL 290
LS + ASLALADAG+ M DL+ + +V + +
Sbjct 121 CCLAFSDFLQWYCIAGDLSVLISCASLALADAGIMMYDLITAVSVSCIGKS--------- 171
Query 291 LLVDPTSDETRSGGPCLTLGVTAETNNIVALQLTGRVDRQTYEKMFEVCLRGCLATAECM 350
L++DP ++E + I L +TG + ++CL E M
Sbjct 172 LMIDPVTEEEGCEDGSFMMTCMPSRYEITQLTITGEWTTPNINEAMQLCLDASSKLGEIM 231
Query 351 K 351
+
Sbjct 232 R 232
> Hs17402904
Length=272
Score = 37.0 bits (84), Expect = 0.068, Method: Compositional matrix adjust.
Identities = 40/108 (37%), Positives = 58/108 (53%), Gaps = 6/108 (5%)
Query 211 EIAAAVRTAAEGIILRRLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLL 270
E+A A++ A E + Y + ++ +S L+L D GS L+A+L AA+LALADAGV M DL+
Sbjct 121 ELALALQEALEPAVRLGRYPRAQLEVSALLLEDGGSALAAALTAAALALADAGVEMYDLV 180
Query 271 PSCTVLLLPQQFPQQQQEPLLLVDPTSDETRSGGPCLTLGVTAETNNI 318
C + L P P L+DPT E LT+ + N +
Sbjct 181 VGCGLSLAPGPAPT------WLLDPTRLEEERAAAGLTVALMPVLNQV 222
> At3g46210
Length=239
Score = 35.0 bits (79), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 17/39 (43%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 231 QTRITISILVLADDGSILSASLIAASLALADAGVAMRDL 269
T ++ I V+ DDGS+L ++ AA AL DAG+ M+ L
Sbjct 99 NTTTSVIIQVVHDDGSLLPCAINAACAALVDAGIPMKHL 137
> SPBC115.01c
Length=226
Score = 33.5 bits (75), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 30/48 (62%), Gaps = 1/48 (2%)
Query 221 EGIILRRLYTQTRITISILVLADDGS-ILSASLIAASLALADAGVAMR 267
E I Y +T I +SI ++ +DG+ L+A + A LAL DAG++++
Sbjct 72 EDAIFLNTYPRTLIQVSIQIIEEDGTDTLAAVINGAVLALLDAGISLK 119
> CE02146
Length=214
Score = 33.1 bits (74), Expect = 0.91, Method: Compositional matrix adjust.
Identities = 19/73 (26%), Positives = 39/73 (53%), Gaps = 9/73 (12%)
Query 227 RLYTQTRITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQ 286
L+ T I++++ + DDGS+ + ++ A AL D G+ + C VL++ +
Sbjct 84 ELFPHTTISVTVHGIQDDGSMGAVAINGACFALLDNGMPFETVF--CGVLIV-------R 134
Query 287 QEPLLLVDPTSDE 299
+ L++DPT+ +
Sbjct 135 VKDELIIDPTAKQ 147
> At5g14580
Length=991
Score = 31.2 bits (69), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 12/69 (17%)
Query 234 ITISILVLADDGSILSASLIAASLALADAGVAMRDLLPSCTVLLLPQQFPQQQQEPLLLV 293
I I+ V++ DGS AS+ S+AL DAG+ +R + +V L+ V
Sbjct 474 IRINSEVMSSDGSTSMASVCGGSMALMDAGIPLRAHVAGVSVGLITD------------V 521
Query 294 DPTSDETRS 302
DP+S E +
Sbjct 522 DPSSGEIKD 530
> Hs9257257
Length=606
Score = 30.0 bits (66), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 30/68 (44%), Gaps = 8/68 (11%)
Query 101 PDGPAIATAGASGSLASCDGR---AVVSLGCTX-----VMALVYGPQPSSNMSTLSTSSS 152
PDG ATA A G + DG+ V +LG + + A+ + P + +S +S
Sbjct 199 PDGNRFATASADGQIYIYDGKTGEKVCALGGSKAHDGGIYAISWSPDSTHLLSASGDKTS 258
Query 153 NTWSVSCT 160
W VS
Sbjct 259 KIWDVSVN 266
> CE23504_1
Length=1180
Score = 30.0 bits (66), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 29/54 (53%), Gaps = 0/54 (0%)
Query 146 TLSTSSSNTWSVSCTDNTSAAMQMMHEGKQGLARGELAVPVSVVCSVGLVDSCV 199
TL +SSN+ SCT TS A+ M G+ G G LA + VV ++G S V
Sbjct 734 TLIATSSNSSQCSCTHLTSFAILMDISGQVGRLSGGLASALDVVSTIGCAISIV 787
> Hs4827056
Length=534
Score = 30.0 bits (66), Expect = 8.9, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 30/68 (44%), Gaps = 8/68 (11%)
Query 101 PDGPAIATAGASGSLASCDGR---AVVSLGCTX-----VMALVYGPQPSSNMSTLSTSSS 152
PDG ATA A G + DG+ V +LG + + A+ + P + +S +S
Sbjct 199 PDGNRFATASADGQIYIYDGKTGEKVCALGGSKAHDGGIYAISWSPDSTHLLSASGDKTS 258
Query 153 NTWSVSCT 160
W VS
Sbjct 259 KIWDVSVN 266
Lambda K H
0.319 0.129 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8551706058
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40