bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
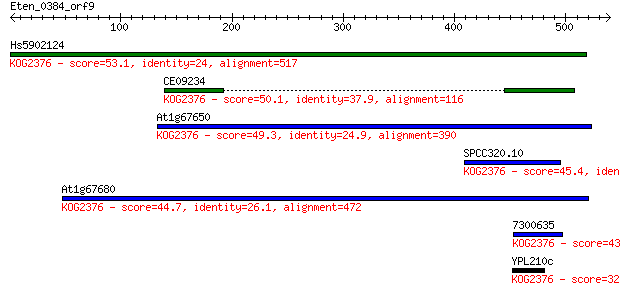
Query= Eten_0384_orf9
Length=539
Score E
Sequences producing significant alignments: (Bits) Value
Hs5902124 53.1 1e-06
CE09234 50.1 1e-05
At1g67650 49.3 2e-05
SPCC320.10 45.4 3e-04
At1g67680 44.7 6e-04
7300635 43.9 8e-04
YPL210c 32.0 4.0
> Hs5902124
Length=671
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 124/530 (23%), Positives = 202/530 (38%), Gaps = 95/530 (17%)
Query 1 LPFNAAALAMQQQQPQAAAALLQRSLELCAAEAGVAVGDALRCIDSGELAGVLVQQAVLQ 60
L +N A + Q Q A +LQ++ +LC D ELA + Q A +
Sbjct 178 LCYNTACALIGQGQLNQAMKILQKAEDLCRRSLSEDT-DGTEEDPQAELAIIHGQMAYIL 236
Query 61 QQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQQQQQQEEQQQQHD 120
Q G A +Y+ ++ + + D G+ AV N +N+ Q +
Sbjct 237 QLQGRTEEALQLYNQII----KLKPTDVGLLAVIANNIIT--INKDQNVFDSK------- 283
Query 121 LDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRFGSSV 180
++ E +E KL+ Q I N+ + + + E+CR++ ASL S+ +
Sbjct 284 -----KKVKLTNAEGVEFKLSKKQLQAIEFNKALLAMYTNQAEQCRKISASLQSQSPEHL 338
Query 181 SLLRLKAAAQFAASRPTKADELLKLLLSSPALSSSSSSNTAAAAATAAQRLQLQICRLAL 240
+ ++AA + TKA ELL+ S N A T AQ L
Sbjct 339 LPVLIQAAQLCREKQHTKAIELLQ------EFSDQHPENAAEIKLTMAQ----------L 382
Query 241 LLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAAAELQQQAGDVEGVL 300
+ QGN +LR +++ L+H ++SA + D++ +
Sbjct 383 KISQGNISKACLILRSIEE--LKHKP--------------GMVSALVTMYSHEEDIDSAI 426
Query 301 ASLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSRWEYAAAQSEAALQRLQQQQQQQQQ 360
+ + Q + A++ L AA R + A+ LQQ +Q +
Sbjct 427 EVFTQAIQWYQNHQPKSPAHLSLIREAANFKLKYGR-------KKEAISDLQQLWKQNPK 479
Query 361 EPAAAAAAAAAELRATLTLAEALARLNLPSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLP 420
+ A +A A+AL++ +LPS+ MSL + DVE
Sbjct 480 DIHTLAQLISAYSLVDPEKAKALSK-HLPSSD----------SMSLKV----DVE----- 519
Query 421 PSSRRAPAAAAAAAAAGKPAADKKKKRK------------KIKYPKGWDPSKPQMPPDPE 468
+ + A GK D + K + K K PK +DP ++ PDPE
Sbjct 520 -ALENSAGATYIRKKGGKVTGDSQPKEQGQGDLKKKKKKKKGKLPKNYDP---KVTPDPE 575
Query 469 RWKPKHERSGYKKMLRKRKECLRG-GAQGAVAPGIAEAVTGFRDSGPTTA 517
RW P ERS Y+ + +K+ G G QGA A +E S P T+
Sbjct 576 RWLPMRERSYYRGRKKGKKKDQIGKGTQGATAGASSELDASKTVSSPPTS 625
> CE09234
Length=694
Score = 50.1 bits (118), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 25/65 (38%), Positives = 41/65 (63%), Gaps = 6/65 (9%)
Query 445 KKRKKIKYPKGWDPSKPQMPPDPERWKPKHERSGYKKMLRKRKECLRGGAQGAVA--PGI 502
K+++KI+ PK ++ + + PDPERW P+ ERS YK+ + R+ + G QG+ + P +
Sbjct 598 KRKRKIRLPKNYNSA---VTPDPERWLPRQERSTYKRKRKNREREIGRGTQGSSSANPNV 654
Query 503 AEAVT 507
E VT
Sbjct 655 -EYVT 658
Score = 37.0 bits (84), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 29/53 (54%), Gaps = 0/53 (0%)
Query 139 KLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRFGSSVSLLRLKAAAQF 191
KLT Q L + +N + LL + E C+R L L ++FGSS + ++A F
Sbjct 349 KLTRRQRLTLMLNNALVLLLSNQREPCKRALEELVAKFGSSKDVALIEATLHF 401
> At1g67650
Length=651
Score = 49.3 bits (116), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 97/401 (24%), Positives = 169/401 (42%), Gaps = 85/401 (21%)
Query 133 TETLERKLTVPQALGIGINRCIGLLKGGRVEECRRVLASLTSRFGSSVSLLRLKAAAQFA 192
++ L+ KL+ I NR + LL ++++ R + A+L F SV L+AA
Sbjct 303 SQELDAKLSHKHKEAIYANRVLLLLHANKMDQARELCATLPGMFPESVIPTLLQAAVLVR 362
Query 193 ASRPTKADELL-----------KLLLSSPALSSSSSSNTAAAAATAAQRLQLQICRLALL 241
++ KA+ELL KL+L + A ++S+S+ AA + ++
Sbjct 363 ENKAAKAEELLGQCAENFPEKSKLVLLARAQIAASASHPHVAAESLSK------------ 410
Query 242 LQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSSREYLALLSAAAELQQQAGDVEGVLA 301
+ D+Q HL A ++ L+++AGD +G A
Sbjct 411 ------------IPDIQ-----HLP--------------ATVATIVALRERAGDNDGATA 439
Query 302 SLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSRWEYAAAQSEAALQRLQQQQQQQQQE 361
LD SA + +T + + L AA +L+ Q+++
Sbjct 440 VLD-----------SAIRWWSDSMTDSNMLRIL--------MPVAAAFKLRHGQEEEASR 480
Query 362 PAAAAAAAAAELRATLTLAEALARLNLPSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLPP 421
A + L LAR+N+ A+ Y + ++ LP L D +++EK
Sbjct 481 LYEEIVKNHNSTDALVGLVTTLARVNVEKAEAY-EKQLKPLP-GLKAVDVDNLEKT---S 535
Query 422 SSRRAPAAAAAAAAAGKPAADKKKKRKKIKYPKGWDPSKPQMPPDPERWKPKHERSGYKK 481
++ +AA+ + + +K K+++K KYPKG+D PDPERW P+ ERS Y+
Sbjct 536 GAKPIEGISAASLSQEEVKKEKVKRKRKPKYPKGFDLENSGPTPDPERWLPRRERSSYRP 595
Query 482 MLRKRKECLRGGAQGAVAPGIAEAVTGFRDSGPTTAKTKAA 522
+ ++ G+QGA+ E P+T+K+K A
Sbjct 596 KRKDKRAAQIRGSQGALTKVKQE-------EAPSTSKSKQA 629
> SPCC320.10
Length=561
Score = 45.4 bits (106), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 46/86 (53%), Gaps = 7/86 (8%)
Query 409 FDSEDVEKQLLPPSSRRAPAAAAAAAAAGKPAADKKKKRKKIKYPKGWDPSKPQMPPDPE 468
D +D+E + +P S+ P + A + + +K+RK PK ++P + PDP+
Sbjct 482 IDVDDIEIRGVPVSAAIGPIKRSVEANSSNSSKKTRKRRKPT--PKSFNP---KATPDPQ 536
Query 469 RWKPKHERSGYKKMLRKRKECLRGGA 494
RW PK +R+ K ++ + + ++GG
Sbjct 537 RWIPKRDRTNVK--IKSKGKSMQGGV 560
> At1g67680
Length=664
Score = 44.7 bits (104), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 123/491 (25%), Positives = 204/491 (41%), Gaps = 109/491 (22%)
Query 48 ELAGVLVQQAVLQQQVGNAAAAEDIYSAVLAAVEQQQTIDAGVAAVAVTNAFAAALNRGQ 107
ELA + VQ A +QQ +G + Y + ++ D AVAV N A +G
Sbjct 240 ELAPIAVQLAYVQQILGQTQESTSSYVDFI----KRNLADEPSLAVAVNNLVAL---KGF 292
Query 108 QQQQQEEQQQQHDLDALIRRATKGATETLERKLTVPQALGIGINRCIGLLKGGRVEECRR 167
+ + ++ DL T ++ L+ KL+ I NR + LL ++++ R
Sbjct 293 KDIS--DGLRKFDLLKDKDSQTFQLSQALDAKLSQKHKEAIYANRVLLLLHANKMDQARE 350
Query 168 VLASLTSRFGSSVSLLRLKAAAQFAASRPTKADELL-----------KLLLSSPALSSSS 216
+ A+L F S+ L+AA ++ KA+ELL KL+L + A ++S
Sbjct 351 LCAALPGMFPESIIPTLLQAAVLVRENKAAKAEELLGQCAEKFPEKSKLVLLARAQIAAS 410
Query 217 SSNTAAAAATAAQRLQLQICRLALLLQQGNRQGFRTLLRDVQQQLLQHLQQQQQQQQGSS 276
+S+ AA + ++ + D+Q HL
Sbjct 411 ASHPHVAAESLSK------------------------IPDIQ-----HLP---------- 431
Query 277 REYLALLSAAAELQQQAGDVEGVLASLDLQRRALEETQGSAAAYVDLCLTAAEIASSLSR 336
A ++ L+++AGD +G A LD S++
Sbjct 432 ----ATVATIVALKERAGDNDGAAAVLD---------------------------SAIKW 460
Query 337 WEYAAAQS--------EAALQRLQQQQQQQQQEPAAAAAAAAAELRATLTLAEALARLNL 388
W + +S EAA +L+ Q+++ A + L LAR+N+
Sbjct 461 WSNSMTESSKLRVLMPEAAAFKLRHGQEEEASRLYEEIVKNHNSTDALVGLVTTLARVNV 520
Query 389 PSAQVYVHRLMRWLPMSLDLFDSEDVEKQLLPPSSRRAPAAAAAAAAAGKPAADKKKKRK 448
A+ Y + ++ LP L D + +EK + AAA+++ K K+K+++
Sbjct 521 EKAESY-EKQLKPLP-GLKAVDVDKLEKTY--GAKPIEGAAASSSQEEVKKEKAKRKRKR 576
Query 449 KIKYPKGWDPSKPQMPPDPERWKPKHERSGYKKMLRKRKECLRGGAQGAVAPGIAEAVTG 508
K KYPKG+DP+ P PPDPERW P+ ERS YK + ++ G+QGAV EA
Sbjct 577 KPKYPKGFDPANPGPPPDPERWLPRRERSSYKPKRKDKRAAQIRGSQGAVTKDKQEA--- 633
Query 509 FRDSGPTTAKT 519
P+T+K+
Sbjct 634 ----APSTSKS 640
> 7300635
Length=650
Score = 43.9 bits (102), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 19/44 (43%), Positives = 28/44 (63%), Gaps = 11/44 (25%)
Query 453 PKGWDPSKPQMPPDPERWKPKHERSGYKKMLRKRKECLRGGAQG 496
PK ++ ++ PDPERW PK+ER+G++K RGGA+G
Sbjct 558 PKNYNA---EVAPDPERWLPKYERTGFRKK--------RGGARG 590
> YPL210c
Length=646
Score = 32.0 bits (71), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 14/29 (48%), Positives = 18/29 (62%), Gaps = 3/29 (10%)
Query 452 YPKGWDPSKPQMPPDPERWKPKHERSGYK 480
+ +G D SK PDPERW P +RS Y+
Sbjct 575 FLQGRDTSKL---PDPERWLPLRDRSTYR 600
Lambda K H
0.314 0.125 0.338
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 14799464076
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40