bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0377_orf2
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
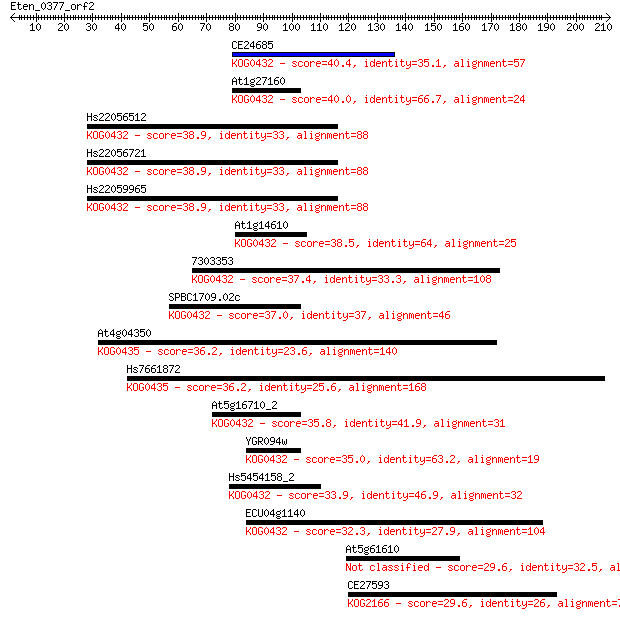
CE24685 40.4 0.002
At1g27160 40.0 0.004
Hs22056512 38.9 0.008
Hs22056721 38.9 0.008
Hs22059965 38.9 0.008
At1g14610 38.5 0.011
7303353 37.4 0.020
SPBC1709.02c 37.0 0.031
At4g04350 36.2 0.045
Hs7661872 36.2 0.051
At5g16710_2 35.8 0.073
YGR094w 35.0 0.096
Hs5454158_2 33.9 0.26
ECU04g1140 32.3 0.67
At5g61610 29.6 4.3
CE27593 29.6 4.6
> CE24685
Length=1050
Score = 40.4 bits (93), Expect = 0.002, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 35/58 (60%), Gaps = 4/58 (6%)
Query 79 CVKTLLTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAASGEWPTTEG-EEFEKEK 135
C+ T L L+ P+ PFI+EELW+R+ + + P +A ++P T+ E+++ EK
Sbjct 833 CIDTGLRLISPLMPFISEELWQRMPRLDDSDYTSPSIIVA---QYPLTQKYEKYQNEK 887
> At1g27160
Length=200
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/24 (66%), Positives = 20/24 (83%), Gaps = 0/24 (0%)
Query 79 CVKTLLTLLHPVAPFITEELWKRL 102
C++T L LLHP P+ITEELW+RL
Sbjct 48 CLETGLRLLHPFMPYITEELWQRL 71
> Hs22056512
Length=1063
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 42/103 (40%), Gaps = 15/103 (14%)
Query 28 EGERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--SCVKTLLT 85
E ER L ++ VT +L ++ + + + R G V SC L
Sbjct 792 ECERGFLTRELSLVTHALHHFWLHNLCDVYLEAVKPVLWHSPRPLGPPQVLFSCADLGLR 851
Query 86 LLHPVAPFITEELWKRL-------------AAEYPAAFAFPGW 115
LL P+ PF+ EELW+RL A YP+A + W
Sbjct 852 LLAPLMPFLAEELWQRLPPRPGCPPAPSISVAPYPSACSLEHW 894
> Hs22056721
Length=1063
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 42/103 (40%), Gaps = 15/103 (14%)
Query 28 EGERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--SCVKTLLT 85
E ER L ++ VT +L ++ + + + R G V SC L
Sbjct 792 ECERGFLTRELSLVTHALHHFWLHNLCDVYLEAVKPVLWHSPRPLGPPQVLFSCADLGLR 851
Query 86 LLHPVAPFITEELWKRL-------------AAEYPAAFAFPGW 115
LL P+ PF+ EELW+RL A YP+A + W
Sbjct 852 LLAPLMPFLAEELWQRLPPRPGCPPAPSISVAPYPSACSLEHW 894
> Hs22059965
Length=694
Score = 38.9 bits (89), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 29/103 (28%), Positives = 42/103 (40%), Gaps = 15/103 (14%)
Query 28 EGERKLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--SCVKTLLT 85
E ER L ++ VT +L ++ + + + R G V SC L
Sbjct 423 ECERGFLTRELSLVTHALHHFWLHNLCDVYLEAVKPVLWHSPRPLGPPQVLFSCADLGLR 482
Query 86 LLHPVAPFITEELWKRL-------------AAEYPAAFAFPGW 115
LL P+ PF+ EELW+RL A YP+A + W
Sbjct 483 LLAPLMPFLAEELWQRLPPRPGCPPAPSISVAPYPSACSLEHW 525
> At1g14610
Length=1108
Score = 38.5 bits (88), Expect = 0.011, Method: Compositional matrix adjust.
Identities = 16/25 (64%), Positives = 20/25 (80%), Gaps = 0/25 (0%)
Query 80 VKTLLTLLHPVAPFITEELWKRLAA 104
++T L LLHP PF+TEELW+RL A
Sbjct 888 LETGLRLLHPFMPFVTEELWQRLPA 912
> 7303353
Length=1049
Score = 37.4 bits (85), Expect = 0.020, Method: Composition-based stats.
Identities = 36/114 (31%), Positives = 55/114 (48%), Gaps = 13/114 (11%)
Query 65 QQQQQRQGGTLSVSCVKTLLTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLA---ASG 121
++QQ TL V C+ L LL P PFITEEL++RL PA P +A ++
Sbjct 831 EEQQTAARRTLYV-CLDYGLRLLSPFMPFITEELYQRLPRANPA----PSICVASYPSNT 885
Query 122 EWPTTEGE---EFEKEKTAENSRKVQVSVQFNGRHRLSVPLSVTEMSTPSDLFE 172
W +T+ E EF +K A R + + + V + T+ S PS++ +
Sbjct 886 SWRSTKIESDVEF-VQKAARIIRSARSDYNLPNKVKTEVYIVCTD-SVPSEILK 937
> SPBC1709.02c
Length=980
Score = 37.0 bits (84), Expect = 0.031, Method: Composition-based stats.
Identities = 17/46 (36%), Positives = 29/46 (63%), Gaps = 3/46 (6%)
Query 57 LMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHPVAPFITEELWKRL 102
L++ +QQ+ +Q TL + + L L+HP P++TEE+W+RL
Sbjct 823 LLSDGTEVQQESAKQ--TL-YTVLDNALRLMHPFMPYVTEEMWQRL 865
> At4g04350
Length=973
Score = 36.2 bits (82), Expect = 0.045, Method: Composition-based stats.
Identities = 33/140 (23%), Positives = 58/140 (41%), Gaps = 24/140 (17%)
Query 32 KLLAPLVANVTRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSVSCVKTLLTLLHPVA 91
+ L +A VT +E + N ++ +M + + G ++ + LL P A
Sbjct 815 RTLHKCIAKVTEEIESTRFNTGISGMMEFVNAAYKWNNQPRGI-----IEPFVLLLSPYA 869
Query 92 PFITEELWKRLAAEYPAAFAFPGWTLAASGEWPTTEGEEFEKEKTAENSRKVQVSVQFNG 151
P + EELW RL +P + A+ + A ++ K T + + VQ NG
Sbjct 870 PHMAEELWSRLG--HPNSLAYESFPKA---------NPDYLKNTT------IVLPVQING 912
Query 152 RHRLSVPLSVTEMSTPSDLF 171
+ R ++ V E + D F
Sbjct 913 KTRGTI--EVEEGCSEDDAF 930
> Hs7661872
Length=903
Score = 36.2 bits (82), Expect = 0.051, Method: Compositional matrix adjust.
Identities = 43/179 (24%), Positives = 72/179 (40%), Gaps = 30/179 (16%)
Query 42 TRSLERMQVNRAVAALMAGSRHLQQQQQRQGGTLSV--------SCVKTLLTLLHPVAPF 93
T E +N A++ LM S L Q Q SV + L+ + P+AP
Sbjct 741 THFTEDFSLNSAISQLMGLSNALSQASQ------SVILHSPEFEDALCALMVMAAPLAPH 794
Query 94 ITEELWKRLAAEYPAAFAFPGWTLAASGE-WPTTEGEEFEKEKTAENSRKVQVSVQFNGR 152
+T E+W LA A W + + WP + E ++ + VQ++V N +
Sbjct 795 VTSEIWAGLALVPRKLCAHYTWDASVLLQAWPAVDPEFLQQPEV------VQMAVLINNK 848
Query 153 H--RLSVPLSVTEMSTPSDLFETVRRFSLVQQRLQLENDRGKALSRVISKPEACLVNFI 209
++ VP V + V F L Q L + +G+++ + P L+NF+
Sbjct 849 ACGKIPVPQQVARDQ------DKVHEFVL-QSELGVRLLQGRSIKKSFLSPRTALINFL 900
> At5g16710_2
Length=830
Score = 35.8 bits (81), Expect = 0.073, Method: Compositional matrix adjust.
Identities = 13/31 (41%), Positives = 20/31 (64%), Gaps = 0/31 (0%)
Query 72 GGTLSVSCVKTLLTLLHPVAPFITEELWKRL 102
G + + + +L LLHP PF+TE+LW+ L
Sbjct 616 GDAVLLYVFENILKLLHPFMPFVTEDLWQAL 646
> YGR094w
Length=1104
Score = 35.0 bits (79), Expect = 0.096, Method: Composition-based stats.
Identities = 12/19 (63%), Positives = 16/19 (84%), Gaps = 0/19 (0%)
Query 84 LTLLHPVAPFITEELWKRL 102
L L+HP PFI+EE+W+RL
Sbjct 897 LKLIHPFMPFISEEMWQRL 915
> Hs5454158_2
Length=985
Score = 33.9 bits (76), Expect = 0.26, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 20/32 (62%), Gaps = 0/32 (0%)
Query 78 SCVKTLLTLLHPVAPFITEELWKRLAAEYPAA 109
+C+ L LL P PF+TEEL++RL P A
Sbjct 772 TCLDVGLRLLSPFMPFVTEELFQRLPRRMPQA 803
> ECU04g1140
Length=921
Score = 32.3 bits (72), Expect = 0.67, Method: Composition-based stats.
Identities = 29/108 (26%), Positives = 54/108 (50%), Gaps = 20/108 (18%)
Query 84 LTLLHPVAPFITEELWKRLAAEYPAAFAFPGWTLAASGEWPTTEGEEFEKE--KTAENSR 141
+ +LHP PFITEE++ F G +++ S +P T+G E E + T + +R
Sbjct 806 MKMLHPFMPFITEEVFSNY---------FNG-SISVS-PYPETDGSEHESKFSVTLQITR 854
Query 142 KVQVSVQFNGRHRLSVPLSVTEMSTPSDLFETVRRF--SLVQQRLQLE 187
++ + NG + +V E++ D+ RF SL ++ ++L+
Sbjct 855 HIRAKAESNGWSK-----AVVEIAPGGDVNHADLRFIRSLCRKIVELK 897
> At5g61610
Length=225
Score = 29.6 bits (65), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 13/40 (32%), Positives = 21/40 (52%), Gaps = 0/40 (0%)
Query 119 ASGEWPTTEGEEFEKEKTAENSRKVQVSVQFNGRHRLSVP 158
A G+ P E + +K+K AE + V+ + G LS+P
Sbjct 179 AEGDKPVEEDKPPQKDKPAEGDKHVEEDMPLGGVEHLSIP 218
> CE27593
Length=777
Score = 29.6 bits (65), Expect = 4.6, Method: Composition-based stats.
Identities = 19/81 (23%), Positives = 41/81 (50%), Gaps = 8/81 (9%)
Query 120 SGEWPTTEGEEFEKEKTAE-----NSRKVQVSVQFNGRHRLSVPLSVTEMSTPS-DLFET 173
+G P++ GE ++ K N+ ++ + +QFN +R+S + E+ P +L
Sbjct 580 NGPGPSSSGESMKERKPEHKILQVNTHQMIILLQFNHHNRISCQQLMDELKIPERELKRN 639
Query 174 VRRFSL--VQQRLQLENDRGK 192
++ +L QR+ + ++GK
Sbjct 640 LQSLALGKASQRILVRKNKGK 660
Lambda K H
0.316 0.130 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3825978242
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40