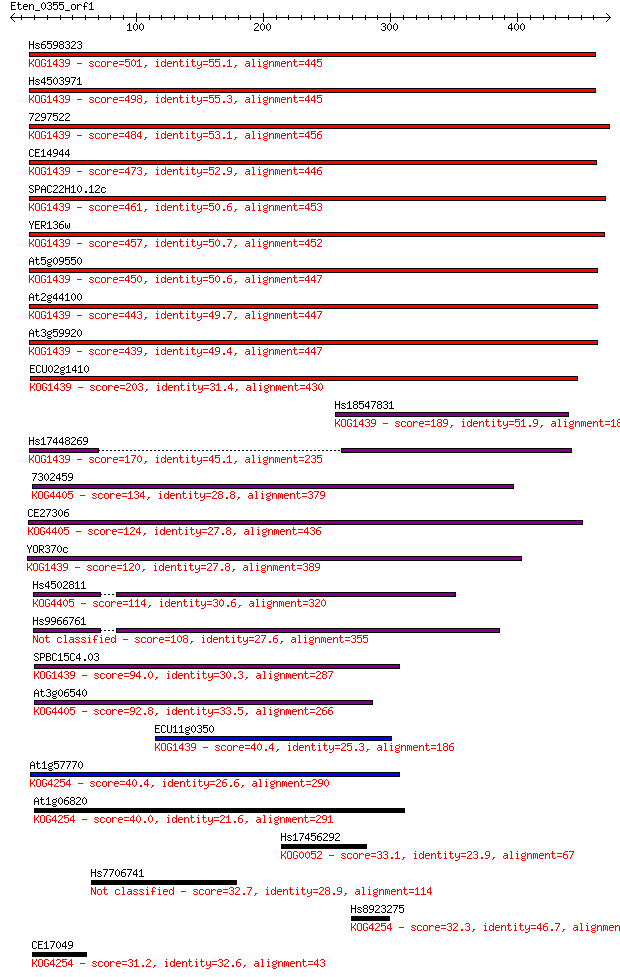
bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0355_orf1
Length=472
Score E
Sequences producing significant alignments: (Bits) Value
Hs6598323 501 2e-141
Hs4503971 498 1e-140
7297522 484 3e-136
CE14944 473 3e-133
SPAC22H10.12c 461 2e-129
YER136w 457 2e-128
At5g09550 450 3e-126
At2g44100 443 3e-124
At3g59920 439 9e-123
ECU02g1410 203 7e-52
Hs18547831 189 1e-47
Hs17448269 170 7e-42
7302459 134 4e-31
CE27306 124 6e-28
YOR370c 120 8e-27
Hs4502811 114 3e-25
Hs9966761 108 2e-23
SPBC15C4.03 94.0 6e-19
At3g06540 92.8 1e-18
ECU11g0350 40.4 0.008
At1g57770 40.4 0.009
At1g06820 40.0 0.012
Hs17456292 33.1 1.5
Hs7706741 32.7 1.6
Hs8923275 32.3 2.3
CE17049 31.2 4.6
> Hs6598323
Length=445
Score = 501 bits (1289), Expect = 2e-141, Method: Compositional matrix adjust.
Identities = 245/446 (54%), Positives = 325/446 (72%), Gaps = 16/446 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLS-LTNLYNKFKPGT 74
M+EEYDV+V GTGL ECILSG++S GKKVLH+DRN YYGGE AS++ L +LY +FK
Sbjct 1 MNEEYDVIVLGTGLTECILSGIMSVNGKKVLHMDRNPYYGGESASITPLEDLYKRFKIPG 60
Query 75 SPPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFS 134
SPP G R+WNVDLIPKF+MA G+LVK+LL T+VTRYL+++V EG+ VY+ K
Sbjct 61 SPPESMGRGRDWNVDLIPKFLMANGQLVKMLLYTEVTRYLDFKVTEGSFVYKGGK----- 115
Query 135 GEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQV 194
I+KVP+TE+EAL S LM L EK R R F Y A P T++G DPK+ +M V
Sbjct 116 ----IYKVPSTEAEALASSLMGLFEKRRFRKFLVYVANFDEKDPRTFEGIDPKKTTMRDV 171
Query 195 YKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYGL 254
YK F L + IDF GHA+ALY DDYL QP ET+ RIKLY S++RYG+SP++YP+YGL
Sbjct 172 YKKFDLGQDVIDFTGHALALYRTDDYLDQPCYETINRIKLYSESLARYGKSPYLYPLYGL 231
Query 255 GGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAE 314
G LP+GF+RL A+ GGTYMLN P++ ++ + GKV GVK+ +G++ARCK ++CDPSY+ +
Sbjct 232 GELPQGFARLSAIYGGTYMLNKPIEEIIV-QNGKVIGVKS-EGEIARCKQLICDPSYVKD 289
Query 315 QFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICL 374
R++ G+VIR ICIL P+ +TN + SCQIIIPQ Q+NR +DIY+ ++S H +
Sbjct 290 ----RVEKVGQVIRVICILSHPIKNTNDANSCQIIIPQNQVNRKSDIYVCMISFAHNVAA 345
Query 375 KGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFD 434
+GK IAIVSTTVET +PEKE+ PAL+LL IE+KFV++SDLL D G +S IF+S ++D
Sbjct 346 QGKYIAIVSTTVETKEPEKEIRPALELLEPIEQKFVSISDLLVPKDLGTESQIFISRTYD 405
Query 435 ATSHFESATQDVLRMWEDMTGEPLDL 460
AT+HFE+ D+ +++ MTG D
Sbjct 406 ATTHFETTCDDIKNIYKRMTGSEFDF 431
> Hs4503971
Length=447
Score = 498 bits (1282), Expect = 1e-140, Method: Compositional matrix adjust.
Identities = 246/446 (55%), Positives = 323/446 (72%), Gaps = 16/446 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLS-LTNLYNKFKPGT 74
MDEEYDV+V GTGL ECILSG++S GKKVLH+DRN YYGGE +S++ L LY +F+
Sbjct 1 MDEEYDVIVLGTGLTECILSGIMSVNGKKVLHMDRNPYYGGESSSITPLEELYKRFQLLE 60
Query 75 SPPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFS 134
PP G R+WNVDLIPKF+MA G+LVK+LL T+VTRYL+++VVEG+ VY+ K
Sbjct 61 GPPESMGRGRDWNVDLIPKFLMANGQLVKMLLYTEVTRYLDFKVVEGSFVYKGGK----- 115
Query 135 GEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQV 194
I+KVP+TE+EAL S LM + EK R R F + A + P+T++G DP+ SM V
Sbjct 116 ----IYKVPSTETEALASNLMGMFEKRRFRKFLVFVANFDENDPKTFEGVDPQTTSMRDV 171
Query 195 YKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYGL 254
Y+ F L + IDF GHA+ALY DDYL QP ET+ RIKLY S++RYG+SP++YP+YGL
Sbjct 172 YRKFDLGQDVIDFTGHALALYRTDDYLDQPCLETVNRIKLYSESLARYGKSPYLYPLYGL 231
Query 255 GGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAE 314
G LP+GF+RL A+ GGTYMLN PVD ++ E GKV GVK+ +G+VARCK ++CDPSYI
Sbjct 232 GELPQGFARLSAIYGGTYMLNKPVDDIIM-ENGKVVGVKS-EGEVARCKQLICDPSYI-- 287
Query 315 QFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICL 374
P R++ G+VIR ICIL P+ +TN + SCQIIIPQ Q+NR +DIY+ ++S H +
Sbjct 288 --PDRVRKAGQVIRIICILSHPIKNTNDANSCQIIIPQNQVNRKSDIYVCMISYAHNVAA 345
Query 375 KGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFD 434
+GK IAI STTVET DPEKE+ PAL+LL I++KFVA+SDL E D+G +S +F S S+D
Sbjct 346 QGKYIAIASTTVETTDPEKEVEPALELLEPIDQKFVAISDLYEPIDDGCESQVFCSCSYD 405
Query 435 ATSHFESATQDVLRMWEDMTGEPLDL 460
AT+HFE+ D+ +++ M G D
Sbjct 406 ATTHFETTCNDIKDIYKRMAGTAFDF 431
> 7297522
Length=443
Score = 484 bits (1245), Expect = 3e-136, Method: Compositional matrix adjust.
Identities = 242/459 (52%), Positives = 324/459 (70%), Gaps = 19/459 (4%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLS-LTNLYNKFKPGT 74
M+EEYD +V GTGLKECILSG+LS +GKKVLH+DRN YYGGE AS++ L L+ ++ G
Sbjct 1 MNEEYDAIVLGTGLKECILSGMLSVSGKKVLHIDRNKYYGGESASITPLEELFQRY--GL 58
Query 75 SPPPE-YGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYF 133
PP E +G R+WNVDLIPKF+MA G+LVK+L+ T VTRYLE++ +EG+ VY+ K
Sbjct 59 EPPGERFGRGRDWNVDLIPKFLMANGQLVKLLIHTGVTRYLEFKSIEGSYVYKGGK---- 114
Query 134 SGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQ 193
I KVP + EAL S LM + EK R R+F Y + D P+TWK FDP + +M+
Sbjct 115 -----IAKVPVDQKEALASDLMGMFEKRRFRNFLIYVQDFREDDPKTWKDFDPTKANMQG 169
Query 194 VYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYG 253
+Y FGL+ NT DF GHA+AL+ D+YL +P T+ RIKLY S++RYG+SP++YP+YG
Sbjct 170 LYDKFGLDKNTQDFTGHALALFRDDEYLNEPAVNTIRRIKLYSDSLARYGKSPYLYPMYG 229
Query 254 LGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIA 313
LG LP+GF+RL A+ GGTYML+ P+D VL E GKV GV++ + +VA+CK V CDPSY+
Sbjct 230 LGELPQGFARLSAIYGGTYMLDKPIDEIVLGEGGKVVGVRSGE-EVAKCKQVYCDPSYV- 287
Query 314 EQFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGIC 373
P +++ G+VIRCICIL PV ST S QIIIPQ+Q+ R +DIY+ +VSS H +
Sbjct 288 ---PEKVRKRGKVIRCICILDHPVASTKDGLSTQIIIPQKQVGRKSDIYVSLVSSTHQVA 344
Query 374 LKGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSF 433
KG + +VSTTVET++PE E+ P L LL I +KFV +SD LE D+G +S IF+S S+
Sbjct 345 AKGWFVGMVSTTVETENPEVEIKPGLDLLEPIAQKFVTISDYLEPIDDGSESQIFISESY 404
Query 434 DATSHFESATQDVLRMWEDMTGEPLDLS-VKAEPEDLQE 471
DAT+HFE+ DVL +++ TGE D S +K E D ++
Sbjct 405 DATTHFETTCLDVLNIFKRGTGETFDFSKIKHELGDEEQ 443
> CE14944
Length=444
Score = 473 bits (1218), Expect = 3e-133, Method: Compositional matrix adjust.
Identities = 236/448 (52%), Positives = 314/448 (70%), Gaps = 17/448 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLS-LTNLYNKFK-PG 73
MDEEYD +V GTGLKECI+SG+LS +GKKVLH+DRNNYYGGE ASL+ L LY KF P
Sbjct 1 MDEEYDAIVLGTGLKECIISGMLSVSGKKVLHIDRNNYYGGESASLTPLEQLYEKFHGPQ 60
Query 74 TSPPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYF 133
P E G R+WNVDLIPKF+MA G LVK+L+ T VTRYLE++ +E + V + K
Sbjct 61 AKPQQEMGRGRDWNVDLIPKFLMANGPLVKLLIHTGVTRYLEFKSIEASFVVKGGK---- 116
Query 134 SGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQ 193
I+KVPA E EAL + LM + EK R + F + Q +K +TW+G DP +M+Q
Sbjct 117 -----IYKVPADEMEALATSLMGMFEKRRFKKFLVWVQQFDENKEDTWQGLDPHNSTMQQ 171
Query 194 VYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYG 253
VY+ FGL+ NT DF GHA+ALY D++ QP +E+I+LY S++RYG+SP++YP+YG
Sbjct 172 VYEKFGLDENTADFTGHALALYRDDEHKNQPYAPAVEKIRLYSDSLARYGKSPYLYPLYG 231
Query 254 LGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIA 313
LG LP+GF+RL A+ GGTYML+ PVD V+ E GK GVK D ++ R K + CDPSY
Sbjct 232 LGELPQGFARLSAIYGGTYMLDKPVDEIVM-ENGKAIGVKCGD-EIVRGKQIYCDPSYAK 289
Query 314 EQFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGIC 373
+ R+K TG+V+R IC+L P+P+TN + SCQIIIPQ+Q+ RH DIYI S+ + +
Sbjct 290 D----RVKKTGQVVRAICLLNHPIPNTNDAQSCQIIIPQKQVGRHYDIYISCCSNTNMVT 345
Query 374 LKGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSF 433
KG +A+VSTTVET +PE E+ P L+LLG I EKF+ +SD+ E +D G +S IF+S S+
Sbjct 346 PKGWYLAMVSTTVETANPEAEVLPGLQLLGAIAEKFIQISDVYEPSDLGSESQIFISQSY 405
Query 434 DATSHFESATQDVLRMWEDMTGEPLDLS 461
DAT+HFE+ +DVL M+E T + D +
Sbjct 406 DATTHFETTCKDVLNMFERGTTKEFDFT 433
> SPAC22H10.12c
Length=440
Score = 461 bits (1185), Expect = 2e-129, Method: Compositional matrix adjust.
Identities = 229/453 (50%), Positives = 304/453 (67%), Gaps = 14/453 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTS 75
MDEEYDV+V GTGL EC+LSGLLS GKKVLH+DRN+YYG + ASL+LT LY F+PG
Sbjct 1 MDEEYDVIVLGTGLTECVLSGLLSVDGKKVLHIDRNDYYGADSASLNLTQLYALFRPGEQ 60
Query 76 PPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSG 135
P G +R+W VDL+PKF+MA G L IL+ T VTRY+E++ + G+ VY+
Sbjct 61 RPESLGRDRDWCVDLVPKFLMANGDLTNILIYTDVTRYIEFKQIAGSYVYR--------- 111
Query 136 EKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQVY 195
+ I KVP E EAL SPLM L EK R + F + + D P T+K + R SME V+
Sbjct 112 DGRIAKVPGNEMEALKSPLMSLFEKRRAKKFLEWVNNYREDDPSTYKDINIDRDSMESVF 171
Query 196 KYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYGLG 255
K FGL++ T DF+GHA+ALY D YLK+P +ET ERI LY S++++G+SP+IYP+YGLG
Sbjct 172 KKFGLQSGTQDFIGHAMALYLDDAYLKKPARETRERILLYASSIAKFGKSPYIYPLYGLG 231
Query 256 GLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAEQ 315
LP+GF+RL A+ GGTYMLN PVD V + G GV++ D QVA+ K ++ DPSY
Sbjct 232 ELPQGFARLSAIYGGTYMLNQPVDEIVYGDDGVAIGVRSGD-QVAKAKQIIGDPSY---- 286
Query 316 FPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICLK 375
F +++ GR++R ICIL P+P+T+ S QIIIPQ Q+ R +DIYI +SS H +C K
Sbjct 287 FREKVRSVGRLVRAICILNHPIPNTDNLDSVQIIIPQNQVKRKHDIYIAGISSVHNVCPK 346
Query 376 GKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFDA 435
G +AI+ST VET +P E+AP LKLLG + E F V ++ E +G ++S S DA
Sbjct 347 GYYLAIISTIVETANPLSEIAPGLKLLGPVVESFSRVQEIYEPVTDGTKDQCYISKSVDA 406
Query 436 TSHFESATQDVLRMWEDMTGEPLDLSVKAEPED 468
TSHFE+ T DV +++ MTG L L + + ED
Sbjct 407 TSHFETLTCDVRDIYKRMTGTDLVLKQRPKMED 439
> YER136w
Length=451
Score = 457 bits (1176), Expect = 2e-128, Method: Compositional matrix adjust.
Identities = 229/458 (50%), Positives = 315/458 (68%), Gaps = 20/458 (4%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTS 75
+D +YDV+V GTG+ ECILSGLLS GKKVLH+D+ ++YGGE AS++L+ LY KFK
Sbjct 6 IDTDYDVIVLGTGITECILSGLLSVDGKKVLHIDKQDHYGGEAASVTLSQLYEKFKQNPI 65
Query 76 PPPE----YGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAG 131
E +G +R+WNVDLIPKF+MA G+L IL+ T VTRY++++ V G+ V++ K
Sbjct 66 SKEERESKFGKDRDWNVDLIPKFLMANGELTNILIHTDVTRYVDFKQVSGSYVFKQGK-- 123
Query 132 YFSGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSM 191
I+KVPA E EA+ SPLM + EK R + F + + + D T +G D +++M
Sbjct 124 -------IYKVPANEIEAISSPLMGIFEKRRMKKFLEWISSYKEDDLSTHQGLDLDKNTM 176
Query 192 EQVYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPV 251
++VY FGL +T +F+GHA+AL+T DDYL+QP + + ERI LY SV+RYG+SP++YP+
Sbjct 177 DEVYYKFGLGNSTKEFIGHAMALWTNDDYLQQPARPSFERILLYCQSVARYGKSPYLYPM 236
Query 252 YGLGGLPEGFSRLCAVNGGTYMLNAPVDSFVL-DEKGKVCGVKTKDGQVARCKMVVCDPS 310
YGLG LP+GF+RL A+ GGTYML+ P+D + + GK GVKTK G + +V+ DP+
Sbjct 237 YGLGELPQGFARLSAIYGGTYMLDTPIDEVLYKKDTGKFEGVKTKLGTF-KAPLVIADPT 295
Query 311 YIAEQFPSRLKCTG-RVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSP 369
Y FP + K TG RVIR ICIL PVP+T+ + S QIIIPQ QL R +DIY+ +VS
Sbjct 296 Y----FPEKCKSTGQRVIRAICILNHPVPNTSNADSLQIIIPQSQLGRKSDIYVAIVSDA 351
Query 370 HGICLKGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFV 429
H +C KG +AI+ST +ETD P EL PA KLLG IEEKF+ +++L E ++G NI++
Sbjct 352 HNVCSKGHYLAIISTIIETDKPHIELEPAFKLLGPIEEKFMGIAELFEPREDGSKDNIYL 411
Query 430 SNSFDATSHFESATQDVLRMWEDMTGEPLDLSVKAEPE 467
S S+DA+SHFES T DV ++ +TG PL L + E E
Sbjct 412 SRSYDASSHFESMTDDVKDIYFRVTGHPLVLKQRQEQE 449
> At5g09550
Length=445
Score = 450 bits (1157), Expect = 3e-126, Method: Compositional matrix adjust.
Identities = 226/448 (50%), Positives = 303/448 (67%), Gaps = 15/448 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTS 75
MDEEYDV+V GTGLKECILSGLLS G KVLH+DRN+YYGGE +SL+LT L+ +F+ +
Sbjct 1 MDEEYDVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGESSSLNLTQLWKRFRGSDT 60
Query 76 PPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSG 135
P G++RE+NVD+IPKF+MA G LV+ L+ T VT+YL ++ V+G+ VY+ K
Sbjct 61 PEENLGASREYNVDMIPKFIMANGLLVQTLIHTDVTKYLNFKAVDGSFVYKKGK------ 114
Query 136 EKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQVY 195
I+KVPAT+ EAL SPLM L EK R R FF Y P++ +G D + + ++
Sbjct 115 ---IYKVPATDVEALKSPLMGLFEKRRARKFFIYVQDYDEKDPKSHEGLDLSKVTAREII 171
Query 196 KYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRY-GRSPFIYPVYGL 254
+GLE +TIDF+GHA+AL+ DDYL QP + ++RIKLY S++R+ G SP+IYP+YGL
Sbjct 172 SKYGLEDDTIDFIGHALALHNDDDYLDQPAIDFVKRIKLYAESLARFQGGSPYIYPLYGL 231
Query 255 GGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAE 314
G LP+ F+RL AV GGTYMLN P D GK GV T G+ A+CK VVCDPSY++E
Sbjct 232 GELPQAFARLSAVYGGTYMLNKPECKVEFDGSGKAIGV-TSAGETAKCKKVVCDPSYLSE 290
Query 315 QFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICL 374
++K G+V R +CI+ P+P TN + S QII+PQ+QL R +D+Y+ S H +
Sbjct 291 ----KVKKVGKVTRAVCIMSHPIPDTNDAHSVQIILPQKQLGRKSDMYLFCCSYAHNVAP 346
Query 375 KGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFD 434
KGK IA VS ETD+PE+EL P ++LLG +E F D T+ + N F+S ++D
Sbjct 347 KGKYIAFVSAEAETDNPEEELKPGIELLGPTDEIFYHSYDTYVPTNIQEEDNCFISATYD 406
Query 435 ATSHFESATQDVLRMWEDMTGEPLDLSV 462
AT+HFES DVL M+ +TG+ LDLSV
Sbjct 407 ATTHFESTVVDVLDMYTKITGKTLDLSV 434
> At2g44100
Length=445
Score = 443 bits (1140), Expect = 3e-124, Method: Compositional matrix adjust.
Identities = 222/448 (49%), Positives = 304/448 (67%), Gaps = 15/448 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTS 75
MDEEY+V+V GTGLKECILSGLLS G KVLH+DRN+YYGGE SL+L L+ KF+
Sbjct 1 MDEEYEVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGESTSLNLNQLWKKFRGEEK 60
Query 76 PPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSG 135
P GS+R++NVD++PKF+MA GKLV++L+ T VT+YL ++ V+G+ V+ K
Sbjct 61 APAHLGSSRDYNVDMMPKFMMANGKLVRVLIHTDVTKYLSFKAVDGSYVFVQGK------ 114
Query 136 EKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQVY 195
+ KVPAT EAL SPLM + EK R FF+Y + P+T G D +R + + +
Sbjct 115 ---VQKVPATPMEALKSPLMGIFEKRRAGKFFSYVQEYDEKDPKTHDGMDLRRVTTKDLI 171
Query 196 KYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRY-GRSPFIYPVYGL 254
FGL+ +TIDF+GHAVAL+ D++L QP +T+ R+KLY S++R+ G SP+IYP+YGL
Sbjct 172 AKFGLKEDTIDFIGHAVALHCNDNHLHQPAYDTVMRMKLYAESLARFQGGSPYIYPLYGL 231
Query 255 GGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAE 314
G LP+ F+RL AV GGTYMLN P DE+GKV GV T +G+ A+CK VVCDPSY+
Sbjct 232 GELPQAFARLSAVYGGTYMLNKPECKVEFDEEGKVSGV-TSEGETAKCKKVVCDPSYLT- 289
Query 315 QFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICL 374
++++ GRV R I I+ P+P+TN S S Q+I+PQ+QL R +D+Y+ S H +
Sbjct 290 ---NKVRKIGRVARAIAIMSHPIPNTNDSQSVQVILPQKQLGRKSDMYVFCCSYSHNVAP 346
Query 375 KGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFD 434
KGK IA VST ETD+P+ EL P + LLG ++E F + D E + N F+S S+D
Sbjct 347 KGKFIAFVSTDAETDNPQTELQPGIDLLGPVDELFFDIYDRYEPVNEPTLDNCFISTSYD 406
Query 435 ATSHFESATQDVLRMWEDMTGEPLDLSV 462
AT+HF++ DVL M++ +TG+ LDLSV
Sbjct 407 ATTHFDTTVVDVLNMYKLITGKELDLSV 434
> At3g59920
Length=444
Score = 439 bits (1128), Expect = 9e-123, Method: Compositional matrix adjust.
Identities = 221/448 (49%), Positives = 299/448 (66%), Gaps = 15/448 (3%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTS 75
MDEEY+V+V GTGLKECILSGLLS G KVLH+DRN+YYGGE SL+L L+ KF+
Sbjct 1 MDEEYEVIVLGTGLKECILSGLLSVDGVKVLHMDRNDYYGGESTSLNLNQLWKKFRGEEK 60
Query 76 PPPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSG 135
P G++R++NVD++PKF+M GKLV+ L+ T VT+YL ++ V+G+ V+ K
Sbjct 61 APEHLGASRDYNVDMMPKFMMGNGKLVRTLIHTDVTKYLSFKAVDGSYVFVKGK------ 114
Query 136 EKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQVY 195
+ KVPAT EAL S LM + EK R FF++ + P+T G D R + +++
Sbjct 115 ---VQKVPATPMEALKSSLMGIFEKRRAGKFFSFVQEYDEKDPKTHDGMDLTRVTTKELI 171
Query 196 KYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRY-GRSPFIYPVYGL 254
+GL+ NTIDF+GHAVAL+T D +L QP +T+ R+KLY S++R+ G SP+IYP+YGL
Sbjct 172 AKYGLDGNTIDFIGHAVALHTNDQHLDQPAFDTVMRMKLYAESLARFQGTSPYIYPLYGL 231
Query 255 GGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAE 314
G LP+ F+RL AV GGTYMLN P DE GKV GV T +G+ A+CK +VCDPSY+
Sbjct 232 GELPQAFARLSAVYGGTYMLNKPECKVEFDEGGKVIGV-TSEGETAKCKKIVCDPSYL-- 288
Query 315 QFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICL 374
P++++ GRV R I I+ P+P+TN S S Q+IIPQ+QL R +D+Y+ S H +
Sbjct 289 --PNKVRKIGRVARAIAIMSHPIPNTNDSHSVQVIIPQKQLARKSDMYVFCCSYSHNVAP 346
Query 375 KGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFD 434
KGK IA VST ETD+P+ EL P LLG ++E F + D E + N F+S S+D
Sbjct 347 KGKFIAFVSTDAETDNPQTELKPGTDLLGPVDEIFFDMYDRYEPVNEPELDNCFISTSYD 406
Query 435 ATSHFESATQDVLRMWEDMTGEPLDLSV 462
AT+HFE+ DVL M+ +TG+ LDLSV
Sbjct 407 ATTHFETTVADVLNMYTLITGKQLDLSV 434
> ECU02g1410
Length=429
Score = 203 bits (516), Expect = 7e-52, Method: Compositional matrix adjust.
Identities = 135/438 (30%), Positives = 227/438 (51%), Gaps = 31/438 (7%)
Query 17 DEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTSP 76
+ EYD V+ GTGL EC + +L+ K+V+ LDRN YG + A+L T L F+ +
Sbjct 4 EHEYDFVILGTGLVECAVGCILARKNKRVILLDRNPMYGSDFATLRYTELETYFQSPSIA 63
Query 77 PPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSGE 136
P + E+++DL PK +A K++K+L++ + YLE+ + G+ +++
Sbjct 64 PELEVYDTEFSIDLTPKLFLADSKMLKMLVRYGIDEYLEFCRIPGSFLWR---------- 113
Query 137 KYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQ--RQADKPETWKGFDPKRHSMEQV 194
K +H VP E++++ + L+ + +K + FF R+A K + +K D +M +
Sbjct 114 KKLHPVPTNEAQSMTTGLIGIWQKPKVMRFFWNVRDYAREAAKGKAYKFKD----TMREE 169
Query 195 YKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYG---RSPFIYPV 251
+K +GL +++ +GH +AL D YL + +ET ++I Y+ S+ Y SP++YP
Sbjct 170 FKEYGLTEESMELIGHGIALNLDDSYLDRHPKETFDKIVTYVRSIICYENSMESPYLYPR 229
Query 252 YGLGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKG-KVCGVKTKDGQVARCKM--VVCD 308
YGL + +GF+R C GG M+NA + ++EK +V + + V R K ++
Sbjct 230 YGLSEIAQGFARSCCTKGGEIMINAEI--LEVNEKNVEVLVREPVNKNVLRIKAGKIISG 287
Query 309 PSYIAEQFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSS 368
SY F + I G PV T G++S QII + +L R NDI+++V+ S
Sbjct 288 QSY----FQRSTLLYEIIRGICIIRGDPVCVTRGASSAQIIFLKEELKRRNDIFVVVLGS 343
Query 369 PHGICLKGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIF 428
+G +AI+ST ET DPE E+ L+ LG+I FV V + D N+
Sbjct 344 EEKSTPEGYKVAIISTVKETSDPENEIKVVLEKLGDIVRYFVDVKPVYRAEDT---ENVI 400
Query 429 VSNSFDATSHFESATQDV 446
+ D + HFE +++
Sbjct 401 FTKGVDESPHFEDIYEEI 418
> Hs18547831
Length=210
Score = 189 bits (481), Expect = 1e-47, Method: Compositional matrix adjust.
Identities = 95/183 (51%), Positives = 132/183 (72%), Gaps = 6/183 (3%)
Query 257 LPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAEQF 316
LP+GF+RL A+ GGTYML P++ ++ + GKV GVK+ +G++A CK +CDPS + +
Sbjct 33 LPQGFARLSAIYGGTYMLKKPIEEIIV-QNGKVIGVKS-EGEIACCKQFICDPSCVKD-- 88
Query 317 PSRLKCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICLKG 376
R++ G+VIR ICIL P+ +TN + SCQIIIPQ Q+NR +DIY+ ++S H + +G
Sbjct 89 --RVEKVGQVIRVICILSHPIKNTNDANSCQIIIPQNQVNRKSDIYVCMISFAHDVAAQG 146
Query 377 KTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFDAT 436
K IAIVSTTVET +PEKE+ PAL+LL E+KFV+ SDLL D G +S IF+S ++DAT
Sbjct 147 KYIAIVSTTVETKEPEKEIRPALELLEPTEQKFVSTSDLLIPKDLGTESQIFISRTYDAT 206
Query 437 SHF 439
F
Sbjct 207 HVF 209
> Hs17448269
Length=286
Score = 170 bits (430), Expect = 7e-42, Method: Compositional matrix adjust.
Identities = 84/181 (46%), Positives = 127/181 (70%), Gaps = 6/181 (3%)
Query 261 FSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAEQFPSRL 320
F+RL A+ GGTYMLN P++ ++ + GKV G+K+ +G++A CK ++CDPSY+ +Q L
Sbjct 91 FARLSAIYGGTYMLNKPIEEIIVQD-GKVIGIKS-EGEIAHCKQLICDPSYVKDQ----L 144
Query 321 KCTGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICLKGKTIA 380
+ G+VIR ICIL + +T+ SCQIIIPQ Q++R +DI + +SS H + + K IA
Sbjct 145 EKVGQVIRVICILSHSIKNTSDVNSCQIIIPQNQVSRKSDICVCTISSAHNVATQEKYIA 204
Query 381 IVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNIFVSNSFDATSHFE 440
IV+TTVET +PEKE+ +L+LL +FV++S LL D G +S IF+S +++ T+HFE
Sbjct 205 IVTTTVETKEPEKEVISSLELLEPTRWEFVSISGLLVPKDLGTESQIFISRAYNPTTHFE 264
Query 441 S 441
+
Sbjct 265 T 265
Score = 36.6 bits (83), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 22/54 (40%), Positives = 28/54 (51%), Gaps = 0/54 (0%)
Query 16 MDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNK 69
M+EEYDV+V G GL ECILS S K + + E + SL L+ K
Sbjct 1 MNEEYDVIVLGIGLMECILSATEGSFVYKCGQIYKVPSTETEALTSSLIGLFKK 54
> 7302459
Length=511
Score = 134 bits (337), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 109/408 (26%), Positives = 179/408 (43%), Gaps = 46/408 (11%)
Query 18 EEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTSPP 77
E++D+VV GTG E ++ S GK VLHLD N YYG +S S+ L + P
Sbjct 7 EQFDLVVIGTGFTESCIAAAGSRIGKSVLHLDSNEYYGDVWSSFSMDALCARLDQEVEPH 66
Query 78 PEYGSNR---------------EWN------------VDLIPKFVMACGKLVKILLKTKV 110
+ R WN +DL P+ + A G+LV++L+K+ +
Sbjct 67 SALRNARYTWHSMEKESETDAQSWNRDSVLAKSRRFSLDLCPRILYAAGELVQLLIKSNI 126
Query 111 TRYLEWQVVEGTSVYQYQKAGYFSGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYC 170
RY E++ V+ + +GE I VP + S+ + + ++EK F C
Sbjct 127 CRYAEFRAVDHVCMRH-------NGE--IVSVPCSRSDVFNTKTLTIVEKRLLMKFLTAC 177
Query 171 AQRQADKPETWKGFDPKRHSMEQVYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETME 230
DK + + + + + + V A+A+ C +E M+
Sbjct 178 NDYGEDKCNE-DSLEFRGRTFLEYLQAQRVTEKISSCVMQAIAM--CGP--STSFEEGMQ 232
Query 231 RIKLYMYSVSRYGRSPFIYPVYGLGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVC 290
R + ++ S+ RYG +PF++P+YG G LP+ F RLCAV GG Y L VD LD
Sbjct 233 RTQRFLGSLGRYGNTPFLFPMYGCGELPQCFCRLCAVYGGIYCLKRAVDDIALDSNSNEF 292
Query 291 GVKTKDGQVARCKMVVCDPSYIAEQFPSRLKCTGRVIRCICILGAPVPSTNGSTSCQIII 350
+ + G+ R K VV P Y LK + R + I +P+ + + +
Sbjct 293 LLSSA-GKTLRAKNVVSAPGYTPVSKGIELK--PHISRGLFISSSPLGNEELNKGGGGVN 349
Query 351 PQRQLNRH--NDIYIMVVSSPHGICLKGKTIAIVSTTVETDDPEKELA 396
R L+ + +++ +S G C +G I ++T ++DP +LA
Sbjct 350 LLRLLDNEGGREAFLIQLSHYTGACPEGLYIFHLTTPALSEDPASDLA 397
> CE27306
Length=510
Score = 124 bits (310), Expect = 6e-28, Method: Compositional matrix adjust.
Identities = 121/503 (24%), Positives = 210/503 (41%), Gaps = 95/503 (18%)
Query 15 KMDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLY------- 67
K+ E DVVV GTGL E IL+ + AG VLHLDRN YYGG+ +S +++ ++
Sbjct 4 KLPESVDVVVLGTGLPEAILASACARAGLSVLHLDRNEYYGGDWSSFTMSMVHEVTENQV 63
Query 68 ------------------------------------------NKFKPGTSPPPEYGSNRE 85
++ KP + E R
Sbjct 64 KKLDSSEISKLSELLTENEQLIELGNREIVENIEMTWIPRGKDEEKPMKTQLEEASQMRR 123
Query 86 WNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSV-YQYQKAGYFSGEKYIHKVPA 144
+++DL+PK +++ G +V+ L ++V+ Y E+++V + +AG ++ VP
Sbjct 124 FSIDLVPKILLSKGAMVQTLCDSQVSHYAEFKLVNRQLCPTETPEAGI-----TLNPVPC 178
Query 145 TESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGF--DPKRHSMEQVYKYFGLET 202
++ E S + +LEK F +C Q E + + + + G+
Sbjct 179 SKGEIFQSNALSILEKRALMKFITFCTQWSTKDTEEGRKLLAEHADRPFSEFLEQMGVGK 238
Query 203 NTIDFVGHAVALYTCDDYLKQPMQET-MERIKLYMYSVSRYGRSPFIYPVYGLGGLPEGF 261
F+ + + + ++P T M +M SV +G SPF++P+YG G L + F
Sbjct 239 TLQSFIINTIGILQ-----QRPTAMTGMLASCQFMDSVGHFGPSPFLFPLYGCGELSQCF 293
Query 262 SRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKDGQVARCKMVVCDPSYIAEQFPSRLK 321
RL AV G Y L PV + V + GK+ V +G C+ +V P ++ E P+
Sbjct 294 CRLAAVFGSLYCLGRPVQAIV-KKDGKITAV-IANGDRVNCRYIVMSPRFVPETVPA--S 349
Query 322 CTGRVIRCICILGAPVPSTNGSTSCQIII-------PQRQLNRHNDIYIMVVSSPHGICL 374
T ++ R I+ A S + Q+ + P ++R ++ ++P G L
Sbjct 350 STLKIER---IVYATDKSIKEAEKEQLTLLNLASLRPDAAVSRLVEVGFEACTAPKGHFL 406
Query 375 -------KGKTIAIVSTTVETDDPEKELAPALKLLGNIEEKFVAVSDLLERTDNGRDSNI 427
+G+T V T E + E+ P K+ F A S + D N+
Sbjct 407 VHATGTQEGET--SVKTIAEKIFEKNEVEPYWKM------SFTANS---MKFDTAGAENV 455
Query 428 FVSNSFDATSHFESATQDVLRMW 450
V+ DA H+ S ++ +++
Sbjct 456 VVAPPVDANLHYASVVEECRQLF 478
> YOR370c
Length=603
Score = 120 bits (300), Expect = 8e-27, Method: Compositional matrix adjust.
Identities = 108/411 (26%), Positives = 187/411 (45%), Gaps = 40/411 (9%)
Query 14 AKMDEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNL------- 66
A ++ DV++ GTG+ E +L+ L+ G VLH+D+N+YYG A+L++ +
Sbjct 41 ATTPDKVDVLIAGTGMVESVLAAALAWQGSNVLHIDKNDYYGDTSATLTVDQIKRWVNEV 100
Query 67 -------YNKFKPGTSPPPEYG--SNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQ 117
Y K S G S+R++ +DL PK + A L+ IL+K++V +YLE+Q
Sbjct 101 NEGSVSCYKNAKLYVSTLIGSGKYSSRDFGIDLSPKILFAKSDLLSILIKSRVHQYLEFQ 160
Query 118 VVEGTSVYQYQKAGYFSGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADK 177
+ Y+ K+ T+ E +PL+ K F + +A +
Sbjct 161 SLSNFHTYE---------NDCFEKLTNTKQEIFTDQNLPLMTKRNLMKFIKFVLNWEA-Q 210
Query 178 PETWKGFDPKRHSMEQVYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKLYMY 237
E W+ + +R + + + F LE + + ++ L C D L + E ++RI+ Y+
Sbjct 211 TEIWQPY-AERTMSDFLGEKFKLEKPQVFELIFSIGL--CYD-LNVKVPEALQRIRRYLT 266
Query 238 SVSRYGRSPFIYPVY-GLGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKTKD 296
S YG P + Y G G L +GF R AV G TY LN + SF + KV +D
Sbjct 267 SFDVYGPFPALCSKYGGPGELSQGFCRSAAVGGATYKLNEKLVSF--NPTTKVATF--QD 322
Query 297 G-QVARCKMVVCDPSYIAEQFPSRLKCTGRVIRCICILGAPVPS--TNGSTSCQIIIPQR 353
G +V + V+ P+ + + +V R CI+ P G ++ ++ P
Sbjct 323 GSKVEVSEKVIISPTQAPKDSKHVPQQQYQVHRLTCIVENPCTEWFNEGESAAMVVFPPG 382
Query 354 QLNRHND--IYIMVVSSPHGICLKGKTIAIVSTTVETDDPEKELAPALKLL 402
L N + ++ + IC +G + +STT + E ++ AL+ +
Sbjct 383 SLKSGNKEVVQAFILGAGSEICPEGTIVWYLSTTEQGPRAEMDIDAALEAM 433
> Hs4502811
Length=656
Score = 114 bits (286), Expect = 3e-25, Method: Compositional matrix adjust.
Identities = 72/271 (26%), Positives = 138/271 (50%), Gaps = 23/271 (8%)
Query 84 REWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSGEKYIHKVP 143
R +N+DL+ K + + G L+ +L+K+ V+RY+E++ V T + ++ E + +VP
Sbjct 227 RRFNIDLVSKLLYSQGLLIDLLIKSDVSRYVEFKNV--TRILAFR-------EGKVEQVP 277
Query 144 ATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQVYKYFGLETN 203
+ ++ S + ++EK F +C + + P+ ++ F ++ S + K L N
Sbjct 278 CSRADVFNSKELTMVEKRMLMKFLTFCLEYEQ-HPDEYQAF--RQCSFSEYLKTKKLTPN 334
Query 204 TIDFVGHAVALYT---CDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYGLGGLPEG 260
FV H++A+ + C + + K ++ + R+G +PF++P+YG G +P+G
Sbjct 335 LQHFVLHSIAMTSESSCTTI------DGLNATKNFLQCLGRFGNTPFLFPLYGQGEIPQG 388
Query 261 FSRLCAVNGGTYMLNAPVDSFVLD-EKGKVCGVKTKDGQVARCKMVVCDPSYIAEQFPSR 319
F R+CAV GG Y L V FV+D E G+ + GQ K + + SY++E+ S
Sbjct 389 FCRMCAVFGGIYCLRHKVQCFVVDKESGRCKAIIDHFGQRINAKYFIVEDSYLSEETCSN 448
Query 320 LKCTGRVIRCICILGAPVPSTNGSTSCQIII 350
++ ++ R + I + T+ I+I
Sbjct 449 VQ-YKQISRAVLITDQSILKTDLDQQTSILI 478
Score = 57.8 bits (138), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 26/53 (49%), Positives = 36/53 (67%), Gaps = 0/53 (0%)
Query 19 EYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFK 71
E+DVV+ GTGL E IL+ S +G++VLH+D +YYGG AS S + L + K
Sbjct 8 EFDVVIIGTGLPESILAAACSRSGQRVLHIDSRSYYGGNWASFSFSGLLSWLK 60
> Hs9966761
Length=653
Score = 108 bits (271), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 73/303 (24%), Positives = 148/303 (48%), Gaps = 17/303 (5%)
Query 84 REWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSGEKYIHKVP 143
R +N+DL+ K + + G L+ +L+K+ V+RY E++ + T + ++ E + +VP
Sbjct 225 RRFNIDLVSKLLYSRGLLIDLLIKSNVSRYAEFKNI--TRILAFR-------EGRVEQVP 275
Query 144 ATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKPETWKGFDPKRHSMEQVYKYFGLETN 203
+ ++ S + ++EK F +C + + P+ +KG++ + + K L N
Sbjct 276 CSRADVFNSKQLTMVEKRMLMKFLTFCMEYEK-YPDEYKGYE--EITFYEYLKTQKLTPN 332
Query 204 TIDFVGHAVALYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYGLGGLPEGFSR 263
V H++A+ + + ++ K +++ + RYG +PF++P+YG G LP+ F R
Sbjct 333 LQYIVMHSIAMTS---ETASSTIDGLKATKNFLHCLGRYGNTPFLFPLYGQGELPQCFCR 389
Query 264 LCAVNGGTYMLNAPVDSFVLDEKGKVC-GVKTKDGQVARCKMVVCDPSYIAEQFPSRLKC 322
+CAV GG Y L V V+D++ + C + + GQ + + + SY E SR++
Sbjct 390 MCAVFGGIYCLRHSVQCLVVDKESRKCKAIIDQFGQRIISEHFLVEDSYFPENMCSRVQ- 448
Query 323 TGRVIRCICILGAPVPSTNGSTSCQIIIPQRQLNRHNDIYIMVVSSPHGICLKGKTIAIV 382
++ R + I V T+ I+ + + ++ + S C+KG + +
Sbjct 449 YRQISRAVLITDRSVLKTDSDQQISILTVPAEEPGTFAVRVIELCSSTMTCMKGTYLVHL 508
Query 383 STT 385
+ T
Sbjct 509 TCT 511
Score = 56.6 bits (135), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 25/53 (47%), Positives = 36/53 (67%), Gaps = 0/53 (0%)
Query 19 EYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFK 71
E+DV+V GTGL E I++ S +G++VLH+D +YYGG AS S + L + K
Sbjct 8 EFDVIVIGTGLPESIIAAACSRSGRRVLHVDSRSYYGGNWASFSFSGLLSWLK 60
> SPBC15C4.03
Length=459
Score = 94.0 bits (232), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 87/318 (27%), Positives = 143/318 (44%), Gaps = 53/318 (16%)
Query 20 YDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYN---KFKPGTSP 76
YDV++ GT L+ ILS LS A ++VLH+D N++YG SL+L +L K K S
Sbjct 7 YDVIIVGTNLRNSILSAALSWANQRVLHIDENSFYGEIDGSLTLRDLEQINEKIKKVDSS 66
Query 77 P--PEYGSNRE---------WNVDLIPKF-----------VMACGKLVKILLKTKVTRYL 114
+ GS++ N DLIPK + A +LVK+L +TK+ +YL
Sbjct 67 QILNDNGSHKSPLKRFEVQFLNKDLIPKNKGSVIQFHPQEIFASSELVKLLSETKIYKYL 126
Query 115 EWQVVEGTSVYQYQKAGYFSGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCA--- 171
+ + S E++I KVP + ++ + + L K F + +
Sbjct 127 LLKPARSFRLLT-------SNEEWI-KVPESRADIFNNKNLSLASKRIVMRFMKFVSNIA 178
Query 172 -QRQADKPETWKGFDPKRHSMEQVYKYFGLETNTIDFVGHAVALYTCDDYLKQ-PMQETM 229
++ + + W+ P +E+V++ G + ++ C K P ++ +
Sbjct 179 DEQNQNLVKEWES-KPFYKFLEEVFQLSGA-------IEESIIYGLCQSLSKDIPTKDAL 230
Query 230 ERIKLYMYSVSRYGRSPFIYPVYGLGG-LPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGK 288
+ + Y +S YG ++ +YG G L +GF R AV GGT+ML +D +DE
Sbjct 231 DTVLKYFHSFGMYGDYSYLLAMYGTGSELCQGFCRSSAVMGGTFMLGQAIDK--IDESKI 288
Query 289 VCGVKTKDGQVARCKMVV 306
V KDG K +V
Sbjct 289 VL----KDGSTLSAKKIV 302
> At3g06540
Length=536
Score = 92.8 bits (229), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 89/322 (27%), Positives = 139/322 (43%), Gaps = 66/322 (20%)
Query 20 YDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSL---------------- 63
YD++V GTG+ E +L+ SS+G VLHLD N +YG ASLSL
Sbjct 15 YDLIVVGTGVSESVLAAAASSSGSSVLHLDPNPFYGSHFASLSLPDLTSFLHSNSVSPPP 74
Query 64 -----------------TNLYNKFKPGTSPPPEYGS------NREWNVDLI-PKFVMACG 99
+L N+ + + S +R +NVDL P+ V
Sbjct 75 SPSSPPLPPSNNHDFISVDLVNRSLYSSVEISSFESEILEEHSRRFNVDLAGPRVVFCAD 134
Query 100 KLVKILLKTKVTRYLEWQVVEGTSVYQYQKAGYFSGEKYIHKVPATESEALMSPLMPLLE 159
+ + ++LK+ Y+E++ ++ + V G SGE + VP + + + LLE
Sbjct 135 ESINLMLKSGANNYVEFKSIDASFV------GDSSGE--LRNVPDSRAAIFKDKSLTLLE 186
Query 160 KNRCRSFFAYCAQRQADKPETWKGFDPK--RHSMEQVYKYF----GLETNTIDFVGHAVA 213
KN+ FF A E K ME + F L + +A+A
Sbjct 187 KNQLMKFFKLVQSHLASSTEKDDSTTVKISEEDMESPFVDFLSKMRLPPKIKSIILYAIA 246
Query 214 LY--------TCDDYLKQPMQETMERIKLYMYSVSRYGRS--PFIYPVYGLGGLPEGFSR 263
+ TC LK +E ++R+ LY+ S+ R+ + IYP+YG G LP+ F R
Sbjct 247 MLDYDQDNTETCRHLLKT--KEGIDRLALYITSMGRFSNALGALIYPIYGQGELPQAFCR 304
Query 264 LCAVNGGTYMLNAPVDSFVLDE 285
AV G Y+L P+ + +LD+
Sbjct 305 RAAVKGCIYVLRMPITALLLDK 326
> ECU11g0350
Length=363
Score = 40.4 bits (93), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 47/187 (25%), Positives = 79/187 (42%), Gaps = 33/187 (17%)
Query 115 EWQVVEGTSVYQYQKAGYFSGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQ 174
E ++E + + QK Y +++P T++E S + L EKN
Sbjct 90 ETGMMEFLHLIKIQKHFYVDPSGKTYRIPYTKTEIADSDALSLKEKNLVGRLLN------ 143
Query 175 ADKPETWKGFDPKRHSMEQVYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKL 234
+ S+E++Y+ L T + + + V CD + ER+
Sbjct 144 ------------GKLSLEELYE--SLSEKTREVLTNGVV---CDG------RNHKERMAR 180
Query 235 YMYSVSRYGRSPFIYPVYGLGGLPEGFSRLCAVNGGTYMLN-APVDSFVLDEKGKVCGVK 293
YM + +G PF+YP +GL + E FSR ++ G Y+L+ A V S E + +K
Sbjct 181 YMRN---FGDVPFLYPRHGLKEISEMFSRCNSMCGAGYVLDSALVISNKTKETLDMFSLK 237
Query 294 TKDGQVA 300
KDG+
Sbjct 238 QKDGKTV 244
> At1g57770
Length=578
Score = 40.4 bits (93), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 77/321 (23%), Positives = 117/321 (36%), Gaps = 51/321 (15%)
Query 17 DEEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNKFKPGTSP 76
+ E DVVV G+G+ LL+ + V+ L+ +++ GG S + KF G S
Sbjct 50 EPEADVVVIGSGIGGLCCGALLARYDQDVIVLESHDHPGGAAHSFEIKGY--KFDSGPSL 107
Query 77 PPEYGSNREWNVDLIPKFVMACGKLVKILLKTKVTRYLEWQVV--EGTSVYQYQKAGYFS 134
S R + + + + A G+ +Y W V EG + + +F
Sbjct 108 FSGLQS-RGPQANPLAQVLDALGE------SFPCKKYDSWMVYLPEGDFLSRIGPTDFFK 160
Query 135 G-EKYIHKVPATESEALMS-------PL------MPLLEKNRCRSFFAYCAQRQADKPET 180
EKY E E L+ PL +P L + A R A P
Sbjct 161 DLEKYAGPSAVQEWEKLLVGAYGAILPLSSAAMALPPLSIRGDLGVLSTAAARYA--PSL 218
Query 181 WKGF---DPKR--------HSMEQVYKYFGLET----NTIDFVGHAVALYTCDDYLKQPM 225
K F PK ++ L+ N ID + +A D L M
Sbjct 219 LKSFIKMGPKGALGATKLLRPFSEIVDSLELKDPFIRNWIDLLAFLLAGVKSDGILSAEM 278
Query 226 QETMERIKLYMYSVSRYGRSPFIYPVYGLGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDE 285
+YM++ YP+ G G + E R GG L + V++ V+ E
Sbjct 279 --------IYMFAEWYKPGCTLEYPIDGTGAVVEALVRGLEKFGGRLSLKSHVENIVI-E 329
Query 286 KGKVCGVKTKDGQVARCKMVV 306
GK GVK ++GQ R + V
Sbjct 330 NGKAVGVKLRNGQFVRARKAV 350
> At1g06820
Length=595
Score = 40.0 bits (92), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 63/316 (19%), Positives = 126/316 (39%), Gaps = 58/316 (18%)
Query 20 YDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECASLSLTNLYNK----FKPGTS 75
YD +V G+G+ + + L+ +VL L++ GG + Y + F G+S
Sbjct 78 YDAIVIGSGIGGLVAATQLAVKEARVLVLEKYLIPGGS------SGFYERDGYTFDVGSS 131
Query 76 PPPEYGSNREWNVDLIPKFVMACGKLVKIL-------------LKTKVTRYLEWQVVEGT 122
+G + + N++LI + + A G+ ++++ L ++ R + + E T
Sbjct 132 V--MFGFSDKGNLNLITQALKAVGRKMEVIPDPTTVHFHLPNNLSVRIHREYDDFIAELT 189
Query 123 SVYQYQKAGY--FSGE--KYIHKVPATESEALMSPLM---PLLEKN-RCRSFFAYCAQRQ 174
S + ++K G F G+ K + + + E ++L P+ +K C + Y Q
Sbjct 190 SKFPHEKEGILGFYGDCWKIFNSLNSLELKSLEEPIYLFGQFFQKPLECLTLAYYLPQNA 249
Query 175 ADKPETWKGFDPKRHSMEQVYKYFGLETNTIDFVGHAVALYTCDDYLKQPMQETMERIKL 234
+ + + + + F+ + + + L+ PM +
Sbjct 250 G-----------------AIARKYIKDPQLLSFIDAECFIVSTVNALQTPMINARQ---- 288
Query 235 YMYSVSRYGRSPFIYPVYGLGGLPEGFSRLCAVNGGTYMLNAPVDSFVLDEKGKVCGVKT 294
+ YG YPV G+GG+ + + G A V S +LD GK GV+
Sbjct 289 -VLCDRHYGG--INYPVGGVGGIAKSLAEGLVDQGSEIQYKANVKSIILDH-GKAVGVRL 344
Query 295 KDGQVARCKMVVCDPS 310
DG+ K ++ + +
Sbjct 345 ADGREFFAKTIISNAT 360
> Hs17456292
Length=262
Score = 33.1 bits (74), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 16/67 (23%), Positives = 34/67 (50%), Gaps = 0/67 (0%)
Query 214 LYTCDDYLKQPMQETMERIKLYMYSVSRYGRSPFIYPVYGLGGLPEGFSRLCAVNGGTYM 273
+Y C K+ +++ ++ + G+SP +Y + G+G +P+G A+ G +
Sbjct 28 IYKCGGINKRSIKKFEKKAAEMGKGSFKDGKSPDVYKIGGIGTVPDGQVETVALKPGVVI 87
Query 274 LNAPVDS 280
APV++
Sbjct 88 TFAPVNT 94
> Hs7706741
Length=258
Score = 32.7 bits (73), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 33/118 (27%), Positives = 48/118 (40%), Gaps = 7/118 (5%)
Query 65 NLYNKFKPGTSPPPEYG-SNREWNVDLIPKFVMACGKLVKILLKTKVTRYLE-WQV--VE 120
N + P T P PE +NR N +L MA + L + R E W +
Sbjct 78 NQWRYSSPWTKPQPEVPVTNRAANCNLHVPGPMAVNQFSPSLARRASVRPGELWHFSSLA 137
Query 121 GTSVYQYQKAGYFSGEKYIHKVPATESEALMSPLMPLLEKNRCRSFFAYCAQRQADKP 178
GTS + GY H VP + + PL+ LL+++RC + A R+ P
Sbjct 138 GTSSLE---PGYSHPFPARHLVPEPQPDGKREPLLSLLQQDRCLARPQESAARENGNP 192
> Hs8923275
Length=334
Score = 32.3 bits (72), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 14/30 (46%), Positives = 16/30 (53%), Gaps = 0/30 (0%)
Query 269 GGTYMLNAPVDSFVLDEKGKVCGVKTKDGQ 298
GG + A V S +LD GK CGV K G
Sbjct 35 GGAVLTKATVQSVLLDSAGKACGVSVKKGH 64
> CE17049
Length=544
Score = 31.2 bits (69), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 22/43 (51%), Gaps = 0/43 (0%)
Query 18 EEYDVVVCGTGLKECILSGLLSSAGKKVLHLDRNNYYGGECAS 60
+ YD ++ G G + L+ AGKKV L+R + GG +
Sbjct 13 QSYDAIIIGGGHNGLTAAAYLTKAGKKVCVLERRHVVGGAAVT 55
Lambda K H
0.318 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 12547240560
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40