bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0341_orf2
Length=221
Score E
Sequences producing significant alignments: (Bits) Value
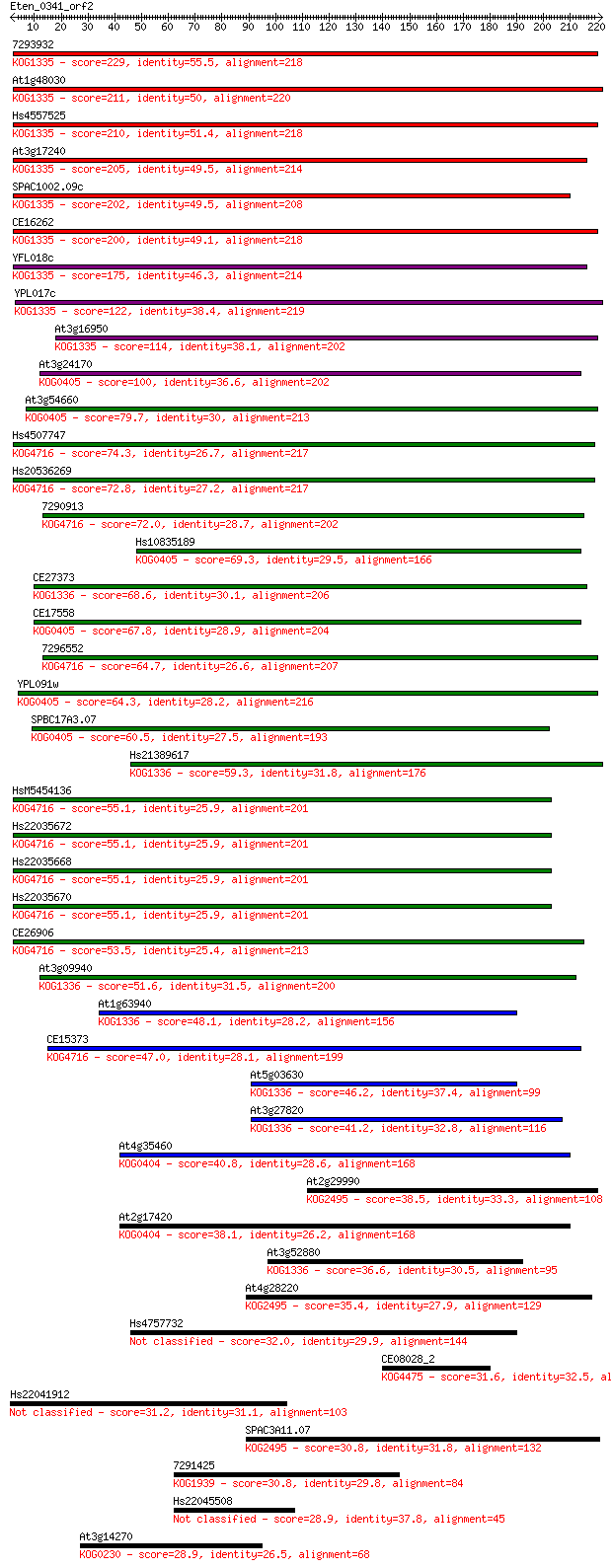
7293932 229 3e-60
At1g48030 211 7e-55
Hs4557525 210 2e-54
At3g17240 205 5e-53
SPAC1002.09c 202 6e-52
CE16262 200 2e-51
YFL018c 175 6e-44
YPL017c 122 5e-28
At3g16950 114 2e-25
At3g24170 100 2e-21
At3g54660 79.7 4e-15
Hs4507747 74.3 2e-13
Hs20536269 72.8 5e-13
7290913 72.0 9e-13
Hs10835189 69.3 5e-12
CE27373 68.6 9e-12
CE17558 67.8 2e-11
7296552 64.7 1e-10
YPL091w 64.3 2e-10
SPBC17A3.07 60.5 2e-09
Hs21389617 59.3 6e-09
HsM5454136 55.1 1e-07
Hs22035672 55.1 1e-07
Hs22035668 55.1 1e-07
Hs22035670 55.1 1e-07
CE26906 53.5 3e-07
At3g09940 51.6 1e-06
At1g63940 48.1 2e-05
CE15373 47.0 3e-05
At5g03630 46.2 5e-05
At3g27820 41.2 0.002
At4g35460 40.8 0.002
At2g29990 38.5 0.010
At2g17420 38.1 0.015
At3g52880 36.6 0.044
At4g28220 35.4 0.095
Hs4757732 32.0 1.1
CE08028_2 31.6 1.4
Hs22041912 31.2 2.0
SPAC3A11.07 30.8 2.1
7291425 30.8 2.2
Hs22045508 28.9 8.8
At3g14270 28.9 8.8
> 7293932
Length=504
Score = 229 bits (584), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 121/221 (54%), Positives = 158/221 (71%), Gaps = 6/221 (2%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V AL+ GI LFK+NKV L G G + P+ V+V+ D G+ + + KN+++ATGSE P
Sbjct 127 VKALTGGIAMLFKKNKVTQLTGFGTIVNPNEVEVKKSD-GSTETVKTKNILIATGSEVTP 185
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G +E+DEE IVSSTGAL L +VPKHLVV+G GVIGLELGSVW LGAEVT +EF D
Sbjct 186 FPG--IEIDEEVIVSSTGALKLAKVPKHLVVIGAGVIGLELGSVWSRLGAEVTAIEFMDT 243
Query 122 II-PALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
I +D E+ + FQK+L +QG+KF GTKV + ++ VT+S+EN KSG+ E+QCD
Sbjct 244 IGGVGIDNEVSKTFQKVLTKQGLKFKLGTKVTAASRSGDNVTVSVENAKSGEKEEIQCDA 303
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
+LV+VGRRPYT+ LGLE +GI D+RGR+ VN Q +VPN
Sbjct 304 LLVSVGRRPYTEGLGLEAVGIVKDDRGRIPVNATFQTVVPN 344
> At1g48030
Length=507
Score = 211 bits (538), Expect = 7e-55, Method: Compositional matrix adjust.
Identities = 110/220 (50%), Positives = 153/220 (69%), Gaps = 4/220 (1%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V L++GIEGLFK+NKV Y+KG G+ P+ V V+ ID GN ++ K++I+ATGS+
Sbjct 132 VKNLTRGIEGLFKKNKVTYVKGYGKFISPNEVSVETIDGGN-TIVKGKHIIVATGSDVKS 190
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L G + +DE+ IVSSTGAL+L VPK L+V+G G IGLE+GSVW LG+EVTVVEF
Sbjct 191 LPG--ITIDEKKIVSSTGALSLSEVPKKLIVIGAGYIGLEMGSVWGRLGSEVTVVEFAGD 248
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVV 181
I+P++D EI + FQ+ LE+Q +KFM TKVV + GV L++E + G+ S ++ DVV
Sbjct 249 IVPSMDGEIRKQFQRSLEKQKMKFMLKTKVVSVDSSSDGVKLTVEPAEGGEQSILEADVV 308
Query 182 LVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNYP 221
LV+ GR P+T L LE++G+ D GR+ VN++ L N P
Sbjct 309 LVSAGRTPFTSGLDLEKIGVETDKAGRILVNDRFL-SNVP 347
> Hs4557525
Length=509
Score = 210 bits (535), Expect = 2e-54, Method: Compositional matrix adjust.
Identities = 112/222 (50%), Positives = 155/222 (69%), Gaps = 7/222 (3%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V AL+ GI LFK+NKV ++ G G++ G + V D G Q++ KN+++ATGSE P
Sbjct 131 VKALTGGIAHLFKQNKVVHVNGYGKITGKNQVTATKADGGT-QVIDTKNILIATGSEVTP 189
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DE+TIVSSTGAL+L +VP+ +VV+G GVIG+ELGSVW+ LGA+VT VEF
Sbjct 190 FPG--ITIDEDTIVSSTGALSLKKVPEKMVVIGAGVIGVELGSVWQRLGADVTAVEFLGH 247
Query 122 IIP-ALDAEIGRAFQKLLERQGIKFMFGTKVVG-SQKADGGVTLSLENVKSGDASEVQCD 179
+ +D EI + FQ++L++QG KF TKV G ++K+DG + +S+E G A + CD
Sbjct 248 VGGVGIDMEISKNFQRILQKQGFKFKLNTKVTGATKKSDGKIDVSIEAASGGKAEVITCD 307
Query 180 VVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+LV +GRRP+TK+LGLEELGI LD RGR+ VN Q +PN
Sbjct 308 VLLVCIGRRPFTKNLGLEELGIELDPRGRIPVNTRFQTKIPN 349
> At3g17240
Length=507
Score = 205 bits (522), Expect = 5e-53, Method: Compositional matrix adjust.
Identities = 106/214 (49%), Positives = 148/214 (69%), Gaps = 3/214 (1%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V L++G+EGLFK+NKV+Y+KG G+ P V V ID G ++ K++I+ATGS+
Sbjct 132 VKNLTRGVEGLFKKNKVNYVKGYGKFLSPSEVSVDTID-GENVVVKGKHIIVATGSDVKS 190
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L G + +DE+ IVSSTGAL+L +PK L+V+G G IGLE+GSVW LG+EVTVVEF
Sbjct 191 LPG--ITIDEKKIVSSTGALSLTEIPKKLIVIGAGYIGLEMGSVWGRLGSEVTVVEFAAD 248
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVV 181
I+PA+D EI + FQ+ LE+Q +KFM TKVVG + GV L +E + G+ + ++ DVV
Sbjct 249 IVPAMDGEIRKQFQRSLEKQKMKFMLKTKVVGVDSSGDGVKLIVEPAEGGEQTTLEADVV 308
Query 182 LVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
LV+ GR P+T L LE++G+ D GR+ VNE+
Sbjct 309 LVSAGRTPFTSGLDLEKIGVETDKGGRILVNERF 342
> SPAC1002.09c
Length=511
Score = 202 bits (513), Expect = 6e-52, Method: Compositional matrix adjust.
Identities = 103/208 (49%), Positives = 142/208 (68%), Gaps = 2/208 (0%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ GIE LFK+NKV+Y KGTG P + V+ ID Q + AKN I+ATGSE P
Sbjct 134 VKSLTSGIEYLFKKNKVEYAKGTGSFIDPQTLSVKGIDGAADQTIKAKNFIIATGSEVKP 193
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DE+ IVSSTGAL+L VPK + V+GGG+IGLE+GSVW LGAEVTVVEF
Sbjct 194 FPG--VTIDEKKIVSSTGALSLSEVPKKMTVLGGGIIGLEMGSVWSRLGAEVTVVEFLPA 251
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVV 181
+ +DA+I +A +++ +QGIKF TK++ ++ V + +EN+K+ Q DV+
Sbjct 252 VGGPMDADISKALSRIISKQGIKFKTSTKLLSAKVNGDSVEVEIENMKNNKRETYQTDVL 311
Query 182 LVAVGRRPYTKDLGLEELGINLDNRGRV 209
LVA+GR PYT+ LGL++LGI++D RV
Sbjct 312 LVAIGRVPYTEGLGLDKLGISMDKSNRV 339
> CE16262
Length=464
Score = 200 bits (509), Expect = 2e-51, Method: Compositional matrix adjust.
Identities = 107/221 (48%), Positives = 152/221 (68%), Gaps = 6/221 (2%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V L+ GI+ LFK NKV +++G + GP+ VQ + D G+ + + A+N+++A+GSE P
Sbjct 117 VKQLTGGIKQLFKANKVGHVEGFATIVGPNTVQAKKND-GSVETINARNILIASGSEVTP 175
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G + +DE+ IVSSTGAL+L +VPK +VV+G GVIGLELGSVW+ LGAEVT VEF
Sbjct 176 FPG--ITIDEKQIVSSTGALSLGQVPKKMVVIGAGVIGLELGSVWQRLGAEVTAVEFLGH 233
Query 122 II-PALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDV 180
+ +D E+ + FQ+ L +QG KF+ TKV+G+ + +T+ +E K G ++CD
Sbjct 234 VGGMGIDGEVSKNFQRSLTKQGFKFLLNTKVMGASQNGSTITVEVEGAKDGKKQTLECDT 293
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
+LV+VGRRPYT+ LGL + I+LDNRGRV VNE Q VP+
Sbjct 294 LLVSVGRRPYTEGLGLSNVQIDLDNRGRVPVNERFQTKVPS 334
> YFL018c
Length=499
Score = 175 bits (444), Expect = 6e-44, Method: Compositional matrix adjust.
Identities = 99/221 (44%), Positives = 146/221 (66%), Gaps = 9/221 (4%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPID-----AGNPQMLMAKNVILATG 56
V L+ GIE LFK+NKV Y KG G ++V P+D +L KN+I+ATG
Sbjct 116 VKQLTGGIELLFKKNKVTYYKGNGSFEDETKIRVTPVDGLEGTVKEDHILDVKNIIVATG 175
Query 57 SEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVV 116
SE P G +E+DEE IVSSTGAL+L +PK L ++GGG+IGLE+GSV+ LG++VTVV
Sbjct 176 SEVTPFPG--IEIDEEKIVSSTGALSLKEIPKRLTIIGGGIIGLEMGSVYSRLGSKVTVV 233
Query 117 EFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGG--VTLSLENVKSGDAS 174
EF +I ++D E+ +A QK L++QG+ F TKV+ +++ D V + +E+ K+
Sbjct 234 EFQPQIGASMDGEVAKATQKFLKKQGLDFKLSTKVISAKRNDDKNVVEIVVEDTKTNKQE 293
Query 175 EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
++ +V+LVAVGRRPY LG E++G+ +D RGR+ +++Q
Sbjct 294 NLEAEVLLVAVGRRPYIAGLGAEKIGLEVDKRGRLVIDDQF 334
> YPL017c
Length=499
Score = 122 bits (306), Expect = 5e-28, Method: Compositional matrix adjust.
Identities = 84/230 (36%), Positives = 122/230 (53%), Gaps = 17/230 (7%)
Query 3 SALSQGIEGL---FKR----NKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILAT 55
SAL IE L +KR N V KGT PH V++ P ++ AK +++AT
Sbjct 102 SALKHNIEELGNVYKRELSKNNVTVYKGTAAFKDPHHVEIAQ-RGMKPFIVEAKYIVVAT 160
Query 56 GSEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTV 115
GS G A +D + I+SS AL+L +P ++GGG IGLE+ ++ NLG+ VT+
Sbjct 161 GSAVIQCPGVA--IDNDKIISSDKALSLDYIPSRFTIMGGGTIGLEIACIFNNLGSRVTI 218
Query 116 VEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKV-VGSQKADGGVTLSLENVKSGDAS 174
VE +I +D E+ A + LL+ QGI F+ T+V + A G + ++L N S
Sbjct 219 VESQSEICQNMDNELASATKTLLQCQGIAFLLDTRVQLAEADAAGQLNITLLNKVSKKTY 278
Query 175 EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRG---RVGVNEQMLVPNYP 221
CDV++V++GRRP K L + +G LD R V V Q L+ YP
Sbjct 279 VHHCDVLMVSIGRRPLLKGLDISSIG--LDERDFVENVDVQTQSLL-KYP 325
> At3g16950
Length=564
Score = 114 bits (284), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 77/209 (36%), Positives = 119/209 (56%), Gaps = 16/209 (7%)
Query 18 VDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAGGALEVDEETIVSS 77
VD L G G + GP Q + G ++ AK++I+ATGS P +EVD +T+++S
Sbjct 192 VDILTGFGSVLGP-----QKVKYGKDNIITAKDIIIATGS--VPFVPKGIEVDGKTVITS 244
Query 78 TGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQK- 136
AL L VP+ + +VG G IGLE V+ LG+EVT +E D+++P D EI + Q+
Sbjct 245 DHALKLESVPEWIAIVGSGYIGLEFSDVYTALGSEVTFIEALDQLMPGFDPEISKLAQRV 304
Query 137 LLERQGIKF---MFGTKVVGSQKADGG-VTLSLENVKSGDASE-VQCDVVLVAVGRRPYT 191
L+ + I + +F +K+ ++ DG V + L + K+ + + ++ D L+A GR P+T
Sbjct 305 LINPRKIDYHTGVFASKITPAR--DGKPVLIELIDAKTKEPKDTLEVDAALIATGRAPFT 362
Query 192 KDLGLEELG-INLDNRGRVGVNEQMLVPN 219
LGLE G I +D R RV + LVPN
Sbjct 363 NGLGLENRGFIPVDERMRVIDGKGTLVPN 391
> At3g24170
Length=499
Score = 100 bits (249), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 74/205 (36%), Positives = 102/205 (49%), Gaps = 16/205 (7%)
Query 12 LFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP-APLAGGALEVD 70
L V +G GR+ GP+ V+V+ ID G AK++++ATGS P G
Sbjct 133 LLANAAVKLYEGEGRVVGPNEVEVRQID-GTKISYTAKHILIATGSRAQKPNIPG----- 186
Query 71 EETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIP--ALDA 128
E ++S AL+L PK +V+GGG I +E S+WR +GA V + F K +P D
Sbjct 187 HELAITSDEALSLEEFPKRAIVLGGGYIAVEFASIWRGMGATVDL--FFRKELPLRGFDD 244
Query 129 EIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRR 188
E+ + LE +G+ T + K D G+ V S E DVVL A GR
Sbjct 245 EMRALVARNLEGRGVNLHPQTSLTQLTKTDQGI-----KVISSHGEEFVADVVLFATGRS 299
Query 189 PYTKDLGLEELGINLDNRGRVGVNE 213
P TK L LE +G+ LD G V V+E
Sbjct 300 PNTKRLNLEAVGVELDQAGAVKVDE 324
> At3g54660
Length=565
Score = 79.7 bits (195), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 64/221 (28%), Positives = 105/221 (47%), Gaps = 24/221 (10%)
Query 7 QGIEGLFK----RNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA-P 61
Q + G++K + V ++G G++ PH V V + ++ +N+++A G P P
Sbjct 186 QRLTGIYKNILSKANVKLIEGRGKVIDPHTVDV------DGKIYTTRNILIAVGGRPFIP 239
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
G +E + S AL LP PK + +VGGG I LE ++ L EV V K
Sbjct 240 DIPG-----KEFAIDSDAALDLPSKPKKIAIVGGGYIALEFAGIFNGLNCEVHVFIRQKK 294
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKA-DGGVTLSLENVKSGDASEVQCDV 180
++ D ++ + + +GI+F KA DG +L K+ +
Sbjct 295 VLRGFDEDVRDFVGEQMSLRGIEFHTEESPEAIIKAGDGSFSL-----KTSKGTVEGFSH 349
Query 181 VLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
V+ A GR+P TK+LGLE +G+ + G + V+E Q VP+
Sbjct 350 VMFATGRKPNTKNLGLENVGVKMAKNGAIEVDEYSQTSVPS 390
> Hs4507747
Length=497
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 58/224 (25%), Positives = 109/224 (48%), Gaps = 12/224 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+ +L+ G + KV Y G+ GPH ++ + G ++ A++ ++ATG P
Sbjct 109 IGSLNWGYRVALREKKVVYENAYGQFIGPHRIKATN-NKGKEKIYSAESFLIATGERPRY 167
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L + D+E +SS +LP P +VVG + LE +G VTV+
Sbjct 168 LG---IPGDKEYCISSDDLFSLPYCPGKTLVVGASYVALECAGFLAGIGLGVTVM-VRSI 223
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENV-KSGDASEV---Q 177
++ D ++ + +E GIKF+ + ++ + G L V +S ++ E+ +
Sbjct 224 LLRGFDQDMANKIGEHMEEHGIKFIRQFVPIKVEQIEAGTPGRLRVVAQSTNSEEIIEGE 283
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVP 218
+ V++A+GR T+ +GLE +G+ ++ + G++ V EQ VP
Sbjct 284 YNTVMLAIGRDACTRKIGLETVGVKINEKTGKIPVTDEEQTNVP 327
> Hs20536269
Length=459
Score = 72.8 bits (177), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 59/224 (26%), Positives = 104/224 (46%), Gaps = 12/224 (5%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
+S+L+ G + V Y+ G H ++ G A ++ATG P
Sbjct 71 ISSLNWGYRLSLREKAVAYVNSYGEFVEHHKIKATN-KKGQETYYTAAQFVIATGERPRY 129
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
L ++ D+E ++S +LP P +VVG + LE G +VTV+
Sbjct 130 LG---IQGDKEYCITSDDLFSLPYCPGKTLVVGASYVALECAGFLAGFGLDVTVM-VRSI 185
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENV-KSGDASEV---Q 177
++ D E+ +E+ G+KF+ V Q+ + G L+ + KS + +E
Sbjct 186 LLRGFDQEMAEKVGSYMEQHGVKFLRKFIPVMVQQLEKGSPGKLKVLAKSTEGTETIEGV 245
Query 178 CDVVLVAVGRRPYTKDLGLEELGINLDNR-GRVGVN--EQMLVP 218
+ VL+A+GR T+ +GLE++G+ ++ + G++ VN EQ VP
Sbjct 246 YNTVLLAIGRDSCTRKIGLEKIGVKINEKSGKIPVNDVEQTNVP 289
> 7290913
Length=491
Score = 72.0 bits (175), Expect = 9e-13, Method: Compositional matrix adjust.
Identities = 58/204 (28%), Positives = 99/204 (48%), Gaps = 10/204 (4%)
Query 13 FKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP-APLAGGALEVDE 71
+ KV+Y+ G G H + + + +G + + A+ ++A G P P GA+E
Sbjct 118 LRDKKVEYINGLGSFVDSHTLLAK-LKSGE-RTITAQTFVIAVGGRPRYPDIPGAVEYG- 174
Query 72 ETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIG 131
++S +L R P +VVG G IGLE + LG E TV+ ++ D ++
Sbjct 175 ---ITSDDLFSLDREPGKTLVVGAGYIGLECAGFLKGLGYEPTVM-VRSIVLRGFDQQMA 230
Query 132 RAFQKLLERQGIKFMFGTKVVGSQKADGGVTL-SLENVKSGDASEVQCDVVLVAVGRRPY 190
+E +GI F+ T + +K D G L +NV++G+ +E D VL A+GR+
Sbjct 231 ELVAASMEERGIPFLRKTVPLSVEKQDDGKLLVKYKNVETGEEAEDVYDTVLWAIGRKGL 290
Query 191 TKDLGLEELGINLDNRGRVGVNEQ 214
DL L G+ + + ++ V+ Q
Sbjct 291 VDDLNLPNAGVTV-QKDKIPVDSQ 313
> Hs10835189
Length=479
Score = 69.3 bits (168), Expect = 5e-12, Method: Compositional matrix adjust.
Identities = 49/175 (28%), Positives = 90/175 (51%), Gaps = 15/175 (8%)
Query 48 AKNVILATGSEPAP-----LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLEL 102
A ++++ATG P+ + G +L + +S G L +P V+VG G I +E+
Sbjct 150 APHILIATGGMPSTPHESQIPGASLGI------TSDGFFQLEELPGRSVIVGAGYIAVEM 203
Query 103 GSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVT 162
+ LG++ +++ DK++ + D+ I + LE G++ + ++V +K G+
Sbjct 204 AGILSALGSKTSLMIRHDKVLRSFDSMISTNCTEELENAGVEVLKFSQVKEVKKTLSGLE 263
Query 163 LSLENVKSGDASEV----QCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE 213
+S+ G + D +L A+GR P TKDL L +LGI D++G + V+E
Sbjct 264 VSMVTAVPGRLPVMTMIPDVDCLLWAIGRVPNTKDLSLNKLGIQTDDKGHIIVDE 318
> CE27373
Length=549
Score = 68.6 bits (166), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 62/217 (28%), Positives = 101/217 (46%), Gaps = 26/217 (11%)
Query 10 EGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLA--GGAL 67
+ ++ V +L T +A H + + + N + ++ +I+ATG L G L
Sbjct 204 DAFYEERNVKFLLKTSVIAVNH--KSREVSLSNGETVVYSKLIIATGGNVRKLQVPGSDL 261
Query 68 E-------VDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
+ V+E I+S+ KH+V VG IG+E+ S A VTV+
Sbjct 262 KNICYLRKVEEANIISNLHP------GKHVVCVGSSFIGMEVASALAEKAASVTVISNTP 315
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGG--VTLSLENVKSGDASEVQC 178
+ +P ++IG+ + E +G+KF VV + D G + LEN K E+
Sbjct 316 EPLPVFGSDIGKGIRLKFEEKGVKFELAANVVALRGNDQGEVSKVILENGK-----ELDV 370
Query 179 DVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQM 215
D+++ +G P TK LE GI LDNRG + V+E+
Sbjct 371 DLLVCGIGVTPATK--FLEGSGIKLDNRGFIEVDEKF 405
> CE17558
Length=473
Score = 67.8 bits (164), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 59/207 (28%), Positives = 97/207 (46%), Gaps = 18/207 (8%)
Query 10 EGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA-PLAGGALE 68
E K + V+Y++G A V+V N KN ++A G +P P GA
Sbjct 115 ESGLKGSSVEYIRGRATFAEDGTVEV------NGAKYRGKNTLIAVGGKPTIPNIKGA-- 166
Query 69 VDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDA 128
E + S G L +P VVVG G I +E+ V NLG++ ++ DK++ D
Sbjct 167 ---EHGIDSDGFFDLEDLPSRTVVVGAGYIAVEIAGVLANLGSDTHLLIRYDKVLRTFDK 223
Query 129 EIGRAFQKLLERQG--IKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVG 186
+ ++ + + T+V K D G+ L+++ +G +VQ ++ A+G
Sbjct 224 MLSDELTADMDEETNPLHLHKNTQVTEVIKGDDGL-LTIKTT-TGVIEKVQ--TLIWAIG 279
Query 187 RRPYTKDLGLEELGINLDNRGRVGVNE 213
R P TK+L LE +G+ D G + V+E
Sbjct 280 RDPLTKELNLERVGVKTDKSGHIIVDE 306
> 7296552
Length=516
Score = 64.7 bits (156), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 55/212 (25%), Positives = 100/212 (47%), Gaps = 12/212 (5%)
Query 13 FKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP-APLAGGALEVDE 71
+ KV+Y+ H ++ + + + ++ V++A G P P GA+E+
Sbjct 141 LRDKKVEYVNSMATFRDSHTIEYVAMPGAEHRQVTSEYVVVAVGGRPRYPDIPGAVELG- 199
Query 72 ETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIG 131
++S + R P +VVG G +GLE + LG E TV+ ++ D ++
Sbjct 200 ---ITSDDIFSYEREPGRTLVVGAGYVGLECACFLKGLGYEPTVM-VRSIVLRGFDRQMS 255
Query 132 RAFQKLLERQGIKFMFGTKVVGS--QKADGGVTLSLENVKSG-DASEVQCDVVLVAVGRR 188
++ +GI F+ GT + + ++ADG + + N + D S+V D VL A+GR+
Sbjct 256 ELLAAMMTERGIPFL-GTTIPKAVERQADGRLLVRYRNTTTQMDGSDV-FDTVLWAIGRK 313
Query 189 PYTKDLGLEELGINL-DNRGRVGVNEQMLVPN 219
+DL L+ G+ D++ V E VP+
Sbjct 314 GLIEDLNLDAAGVKTHDDKIVVDAAEATSVPH 345
> YPL091w
Length=483
Score = 64.3 bits (155), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 61/227 (26%), Positives = 105/227 (46%), Gaps = 23/227 (10%)
Query 4 ALSQGIEGLFKRN----KVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSE- 58
A + G++++N KVD + G R V+VQ D ++ A ++++ATG +
Sbjct 114 AYVHRLNGIYQKNLEKEKVDVVFGWARFNKDGNVEVQKRD-NTTEVYSANHILVATGGKA 172
Query 59 --PAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVV 116
P + G L D S G L PK +VVVG G IG+EL V+ LG+E +V
Sbjct 173 IFPENIPGFELGTD------SDGFFRLEEQPKKVVVVGAGYIGIELAGVFHGLGSETHLV 226
Query 117 EFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK--ADGGVTLSLENVKSGDAS 174
+ ++ D I ++GI +K+V +K + + + + KS D
Sbjct 227 IRGETVLRKFDECIQNTITDHYVKEGINVHKLSKIVKVEKNVETDKLKIHMNDSKSID-- 284
Query 175 EVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNE--QMLVPN 219
D ++ +GR+ + +G E +GI L++ ++ +E VPN
Sbjct 285 --DVDELIWTIGRKSHL-GMGSENVGIKLNSHDQIIADEYQNTNVPN 328
> SPBC17A3.07
Length=464
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 53/200 (26%), Positives = 82/200 (41%), Gaps = 16/200 (8%)
Query 9 IEGLFKRN----KVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEP---AP 61
+ G+++RN V Y+ G P V V D Q+ AK +++A G P +
Sbjct 99 LNGIYERNVNKDGVAYISGHASFVSPTEVAVDMNDGSGTQVFSAKYILIAVGGHPIWPSH 158
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
+ G +D S G L PK + +VG G I +EL V+ LG E + K
Sbjct 159 IPGAEYGID------SDGFFELESQPKRVAIVGAGYIAVELAGVFAALGTETHMFIRQSK 212
Query 122 IIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVV 181
+ D I + GI T + +K + + L + D S D +
Sbjct 213 FLRKFDPIISDGIMDHFQHIGINVH--TNSLEFKKVEKLPSGEL-CIHQQDGSTFNVDTL 269
Query 182 LVAVGRRPYTKDLGLEELGI 201
L A+GR P + L LE+ G+
Sbjct 270 LWAIGRAPKIQGLRLEKAGV 289
> Hs21389617
Length=605
Score = 59.3 bits (142), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 56/180 (31%), Positives = 88/180 (48%), Gaps = 11/180 (6%)
Query 46 LMAKNVILATGSEPAPLAGGALEVDE-ETIVSSTGALALPRVPK--HLVVVGGGVIGLEL 102
L ++LA GS P L+ EV+ TI + A + R+ + ++VVVG G +G+E+
Sbjct 288 LEYSKLLLAPGSSPKTLSCKGKEVENVFTIRTPEDANRVVRLARGRNVVVVGAGFLGMEV 347
Query 103 GSVWRNLGAEVTVVEFCDKIIPALDAE-IGRAFQKLLERQGIKFMFGTKVVGSQKADGGV 161
+ V+VVE + E +GRA K+ E +KF T+V + +G
Sbjct 348 AAYLTEKAHSVSVVELEETPFRRFLGERVGRALMKMFENNRVKFYMQTEVSELRGQEG-- 405
Query 162 TLSLENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPNYP 221
L+ V + V+ DV +V +G P T L + GI LD+RG + VN +M+ N P
Sbjct 406 --KLKEVVLKSSKVVRADVCVVGIGAVPATG--FLRQSGIGLDSRGFIPVN-KMMQTNVP 460
> HsM5454136
Length=524
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 86/206 (41%), Gaps = 13/206 (6%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ G + KV Y H V G +L A ++I+ATG P
Sbjct 136 VKSLNWGHRVQLQDRKVKYFNIKASFVDEHTV-CGVAKGGKEILLSADHIIIATGGRPRY 194
Query 62 LAG--GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFC 119
GALE ++S L P +VVG + LE +G + T++
Sbjct 195 PTHIEGALEYG----ITSDDIFWLKESPGKTLVVGASYVALECAGFLTGIGLDTTIMM-- 248
Query 120 DKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEV 176
+ IP D ++ + + G +F+ G ++ DG + ++ E+ +G
Sbjct 249 -RSIPLRGFDQQMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTG 307
Query 177 QCDVVLVAVGRRPYTKDLGLEELGIN 202
D VL A+GR P T+ L LE+ G++
Sbjct 308 TFDTVLWAIGRVPDTRSLNLEKAGVD 333
> Hs22035672
Length=524
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 86/206 (41%), Gaps = 13/206 (6%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ G + KV Y H V G +L A ++I+ATG P
Sbjct 136 VKSLNWGHRVQLQDRKVKYFNIKASFVDEHTV-CGVAKGGKEILLSADHIIIATGGRPRY 194
Query 62 LAG--GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFC 119
GALE ++S L P +VVG + LE +G + T++
Sbjct 195 PTHIEGALEYG----ITSDDIFWLKESPGKTLVVGASYVALECAGFLTGIGLDTTIMM-- 248
Query 120 DKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEV 176
+ IP D ++ + + G +F+ G ++ DG + ++ E+ +G
Sbjct 249 -RSIPLRGFDQQMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTG 307
Query 177 QCDVVLVAVGRRPYTKDLGLEELGIN 202
D VL A+GR P T+ L LE+ G++
Sbjct 308 TFDTVLWAIGRVPDTRSLNLEKAGVD 333
> Hs22035668
Length=428
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 86/206 (41%), Gaps = 13/206 (6%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ G + KV Y H V G +L A ++I+ATG P
Sbjct 40 VKSLNWGHRVQLQDRKVKYFNIKASFVDEHTV-CGVAKGGKEILLSADHIIIATGGRPRY 98
Query 62 LAG--GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFC 119
GALE ++S L P +VVG + LE +G + T++
Sbjct 99 PTHIEGALEYG----ITSDDIFWLKESPGKTLVVGASYVALECAGFLTGIGLDTTIMM-- 152
Query 120 DKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEV 176
+ IP D ++ + + G +F+ G ++ DG + ++ E+ +G
Sbjct 153 -RSIPLRGFDQQMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTG 211
Query 177 QCDVVLVAVGRRPYTKDLGLEELGIN 202
D VL A+GR P T+ L LE+ G++
Sbjct 212 TFDTVLWAIGRVPDTRSLNLEKAGVD 237
> Hs22035670
Length=494
Score = 55.1 bits (131), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 52/206 (25%), Positives = 86/206 (41%), Gaps = 13/206 (6%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAP 61
V +L+ G + KV Y H V G +L A ++I+ATG P
Sbjct 106 VKSLNWGHRVQLQDRKVKYFNIKASFVDEHTV-CGVAKGGKEILLSADHIIIATGGRPRY 164
Query 62 LAG--GALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFC 119
GALE ++S L P +VVG + LE +G + T++
Sbjct 165 PTHIEGALEYG----ITSDDIFWLKESPGKTLVVGASYVALECAGFLTGIGLDTTIMM-- 218
Query 120 DKIIP--ALDAEIGRAFQKLLERQGIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEV 176
+ IP D ++ + + G +F+ G ++ DG + ++ E+ +G
Sbjct 219 -RSIPLRGFDQQMSSMVIEHMASHGTRFLRGCAPSRVRRLPDGQLQVTWEDSTTGKEDTG 277
Query 177 QCDVVLVAVGRRPYTKDLGLEELGIN 202
D VL A+GR P T+ L LE+ G++
Sbjct 278 TFDTVLWAIGRVPDTRSLNLEKAGVD 303
> CE26906
Length=665
Score = 53.5 bits (127), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 54/225 (24%), Positives = 92/225 (40%), Gaps = 18/225 (8%)
Query 2 VSALSQGIEGLFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPA- 60
+++L+ G + V Y+ G GP + + L A +++TG P
Sbjct 270 IASLNWGYRVQLREKTVTYINSYGEFTGPFEISATN-KKKKVEKLTADRFLISTGLRPKY 328
Query 61 PLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCD 120
P G +E ++S LP P + VG + LE G +VTV+
Sbjct 329 PEIPGV----KEYTITSDDLFQLPYSPGKTLCVGASYVSLECAGFLHGFGFDVTVM-VRS 383
Query 121 KIIPALDAEIGRAFQKLLERQGIKFMFGTKVVGSQ---KADGG-----VTLSLENVKSGD 172
++ D ++ +K + G+KF G Q K D V +N ++G+
Sbjct 384 ILLRGFDQDMAERIRKHMIAYGMKFEAGVPTRIEQIDEKTDEKAGKYRVFWPKKNEETGE 443
Query 173 ASEV--QCDVVLVAVGRRPYTKDLGLEELGINLDNRGRV-GVNEQ 214
EV + + +L+A+GR T D+GL +G+ +V G EQ
Sbjct 444 MQEVSEEYNTILMAIGREAVTDDVGLTTIGVERAKSKKVLGRREQ 488
> At3g09940
Length=441
Score = 51.6 bits (122), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 63/217 (29%), Positives = 97/217 (44%), Gaps = 24/217 (11%)
Query 12 LFKRNKVDYLKGTGRLAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAG-GALEVD 70
+K +D + GT + A + D G ++ + +++ATGS L+ G E D
Sbjct 82 WYKEKGIDLIVGTEIVKADLASKTLVSDDG--KIYKYQTLLIATGSTNIRLSEIGVQEAD 139
Query 71 EETIV-------SSTGALALPRVPKH--LVVVGGGVIGLELGSVWRNLGAEVTVVEFCDK 121
+ I S ALA+ + V++GGG +GLE+ S R EVT+V F +
Sbjct 140 VKNIFYLREIEDSDELALAMELYVQRGKAVIIGGGFLGLEISSALRANNHEVTMV-FPEP 198
Query 122 II--PALDAEIGRAFQKLLERQGIKFMFGTKVVG-SQKADGGVTLSLENVKSGDASEVQC 178
+ AEI ++ +GIK + GT G S +DG VT VK D ++
Sbjct 199 WLVHRFFTAEIASFYESYYANKGIKIIKGTVATGFSTNSDGEVT----EVKLEDGRTLEA 254
Query 179 DVVLVAVGRRPYTK----DLGLEELGINLDNRGRVGV 211
++V+ VG RP T L E+ GI D + V
Sbjct 255 NIVVAGVGARPATSLFKGQLEEEKGGIKTDGFFKTSV 291
> At1g63940
Length=493
Score = 48.1 bits (113), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 44/169 (26%), Positives = 82/169 (48%), Gaps = 25/169 (14%)
Query 34 QVQPIDAGNPQMLMAKNVILATGSEPAPLA---GGALE--------VDEETIVSSTGALA 82
Q DAG + L ++I+ATG + GG L D +++++S G
Sbjct 160 QTLTTDAG--KQLKYGSLIIATGCTASRFPDKIGGHLPGVHYIREVADADSLIASLGKA- 216
Query 83 LPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPAL-DAEIGRAFQKLLERQ 141
K +V+VGGG IG+E+ + + T+V D+++ L + + +++L +
Sbjct 217 -----KKIVIVGGGYIGMEVAAAAVAWNLDTTIVFPEDQLLQRLFTPSLAQKYEELYRQN 271
Query 142 GIKFMFGTKVVGSQK-ADGGVTLSLENVKSGDASEVQCDVVLVAVGRRP 189
G+KF+ G + + +DG V+ VK D S ++ D V++ +G +P
Sbjct 272 GVKFVKGASINNLEAGSDGRVS----AVKLADGSTIEADTVVIGIGAKP 316
> CE15373
Length=503
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 56/209 (26%), Positives = 86/209 (41%), Gaps = 27/209 (12%)
Query 15 RNKVDYLKGTGRLAGPHAVQVQPIDAGNPQ-MLMAKNVILATGSEPA-PLAGGALEVDEE 72
+ K++Y + + D + L A NV+++TG P P GA E
Sbjct 132 QKKINYFNAYAEFVDKDKIVITGTDKNKTKNFLSAPNVVISTGLRPKYPNIPGA-----E 186
Query 73 TIVSSTGALALPRVPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIP--ALDAEI 130
++S L VP ++VGGG + LE L A VE + IP D +
Sbjct 187 LGITSDDLFTLASVPGKTLIVGGGYVALECAGF---LSAFNQNVEVLVRSIPLKGFDRDC 243
Query 131 GRAFQKLLERQGIKFMFGTKV-----VGSQKADGGVTLSLENVKSGDASEVQCDVVLVAV 185
+ L+ G+K +V VGS+K VT + G+ + D V+ A
Sbjct 244 VHFVMEHLKTTGVKVKEHVEVERVEAVGSKKK---VTFT------GNGGVEEYDTVIWAA 294
Query 186 GRRPYTKDLGLEELGINLDNR-GRVGVNE 213
GR P K L L+ G+ D R G++ +E
Sbjct 295 GRVPNLKSLNLDNAGVRTDKRSGKILADE 323
> At5g03630
Length=437
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 37/101 (36%), Positives = 53/101 (52%), Gaps = 6/101 (5%)
Query 91 VVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPAL-DAEIGRAFQKLLERQGIKFMFGT 149
VVVGGG IGLELG+ + +VT+V +P L A I ++ +GI + GT
Sbjct 170 VVVGGGYIGLELGAALKANNLDVTMVYPEPWCMPRLFTAGIASFYEGYYANKGINIVKGT 229
Query 150 KVVG-SQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRP 189
G + ++G VT VK D ++ D+V+V VG RP
Sbjct 230 VASGFTTNSNGEVT----EVKLKDGRTLEADIVIVGVGGRP 266
> At3g27820
Length=488
Score = 41.2 bits (95), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 38/122 (31%), Positives = 63/122 (51%), Gaps = 10/122 (8%)
Query 91 VVVGGGVIGLELGSVWRNLGAEVTVVEFCDKIIPA--LDAEIGRAFQKLLERQGIKFMFG 148
VV+GGG IG+E + VT+V F + A +I ++ +G+KF+ G
Sbjct 166 VVIGGGYIGMECAASLVINKINVTMV-FPEAHCMARLFTPKIASLYEDYYRAKGVKFIKG 224
Query 149 TKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYTK----DLGLEELGINLD 204
T V+ S + D ++ N+K D S + D+V+V +G RP T L +E+ GI ++
Sbjct 225 T-VLTSFEFDSNKKVTAVNLK--DGSHLPADLVVVGIGIRPNTSLFEGQLTIEKGGIKVN 281
Query 205 NR 206
+R
Sbjct 282 SR 283
> At4g35460
Length=333
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 48/181 (26%), Positives = 80/181 (44%), Gaps = 24/181 (13%)
Query 42 NPQMLMAKNVILATGSEPAPLA--------GGALEVDEETIVSSTGALALPRVPKHLVVV 93
+ + ++A VILATG+ L+ GG GA + R K L V+
Sbjct 107 DSKAILADAVILATGAVAKRLSFVGSGEASGGFWNRGISACAVCDGAAPIFR-NKPLAVI 165
Query 94 GGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVV- 152
GGG +E + G++V ++ D + + Q+ L I ++ + VV
Sbjct 166 GGGDSAMEEANFLTKYGSKVYIIHRRDAFRASKIMQ-----QRALSNPKIDVIWNSSVVE 220
Query 153 ----GSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGR 208
G + GG L ++NV +GD S+++ + A+G P TK L + G+ LD+ G
Sbjct 221 AYGDGERDVLGG--LKVKNVVTGDVSDLKVSGLFFAIGHEPATKFL---DGGVELDSDGY 275
Query 209 V 209
V
Sbjct 276 V 276
> At2g29990
Length=508
Score = 38.5 bits (88), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 36/111 (32%), Positives = 58/111 (52%), Gaps = 20/111 (18%)
Query 112 EVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFG-TKVVGSQKADGGVTLSLENVKS 170
VT++E D I+ + D + R K L + G++F+ G K V SQK L L+
Sbjct 278 HVTLIEARD-ILSSFDDRLRRYAIKQLNKSGVRFVRGIVKDVQSQK------LILD---- 326
Query 171 GDASEVQCDVVLVA--VGRRPYTKDLGLEELGINLDNRGRVGVNEQMLVPN 219
D +EV +++ + VG P+ + LGL + D GR+G++E M VP+
Sbjct 327 -DGTEVPYGLLVWSTGVGPSPFVRSLGLPK-----DPTGRIGIDEWMRVPS 371
> At2g17420
Length=383
Score = 38.1 bits (87), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 44/179 (24%), Positives = 80/179 (44%), Gaps = 20/179 (11%)
Query 42 NPQMLMAKNVILATGSEPAPLA--------GGALEVDEETIVSSTGALALPRVPKHLVVV 93
+ + ++A +VI++TG+ L+ GG GA + R K LVV+
Sbjct 157 DSRTVLADSVIISTGAVAKRLSFTGSGEGNGGFWNRGISACAVCDGAAPIFR-NKPLVVI 215
Query 94 GGGVIGLELGSVWRNLGAEVTVVEFCDKIIPALDAEIGRAFQKLLERQGIKFMFGTKVVG 153
GGG +E + G++V ++ D + + Q+ L I+ ++ + VV
Sbjct 216 GGGDSAMEEANFLTKYGSKVYIIHRRDTFRASKIMQ-----QRALSNPKIEVIWNSAVVE 270
Query 154 SQKADGGVTL---SLENVKSGDASEVQCDVVLVAVGRRPYTKDLGLEELGINLDNRGRV 209
+ + G L ++NV +GD S+++ + A+G P TK L + + LD G V
Sbjct 271 AYGDENGRVLGGLKVKNVVTGDVSDLKVSGLFFAIGHEPATKFLDGQ---LELDEDGYV 326
> At3g52880
Length=434
Score = 36.6 bits (83), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 29/96 (30%), Positives = 47/96 (48%), Gaps = 4/96 (4%)
Query 97 VIGLELGSVWRNLGAEVTVVEFCDKIIPAL-DAEIGRAFQKLLERQGIKFMFGTKVVGSQ 155
IGLEL +V R +VT+V +P L A+I ++ +G+K + GT G
Sbjct 173 YIGLELSAVLRINNLDVTMVFPEPWCMPRLFTADIAAFYETYYTNKGVKIIKGTVASGFT 232
Query 156 KADGGVTLSLENVKSGDASEVQCDVVLVAVGRRPYT 191
G ++ V+ D ++ D+V+V VG +P T
Sbjct 233 AQPNG---EVKEVQLKDGRTLEADIVIVGVGAKPLT 265
> At4g28220
Length=559
Score = 35.4 bits (80), Expect = 0.095, Method: Compositional matrix adjust.
Identities = 36/145 (24%), Positives = 63/145 (43%), Gaps = 23/145 (15%)
Query 89 HLVVVGGGVIGLELGSVWRNLGAE--------------VTVVEFCDKIIPALDAEIGRAF 134
H V+VGGG G+E + + E +T+++ D I+ D I
Sbjct 216 HFVIVGGGPTGVEFAAELHDFIIEDITKIYPSVKELVKITLIQSGDHILNTFDERISSFA 275
Query 135 QKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVA--VGRRPYTK 192
++ R GI G +V+ D +T+ +++ SG+ + ++L + VG RP
Sbjct 276 EQKFTRDGIDVQTGMRVMSV--TDKDITVKVKS--SGELVSIPHGLILWSTGVGTRPVIS 331
Query 193 DLGLEELGINLDNRGRVGVNEQMLV 217
D +E++G R V NE + V
Sbjct 332 DF-MEQVGQG--GRRAVATNEWLQV 353
> Hs4757732
Length=613
Score = 32.0 bits (71), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 43/161 (26%), Positives = 68/161 (42%), Gaps = 26/161 (16%)
Query 46 LMAKNVILATGSEPAPLAG----GALEVDEETIVSSTGAL----ALPRVPKHLVVVGGGV 97
+ + ++ATG P L+ GA T+ G + R K + ++GGG
Sbjct 251 ITYEKCLIATGGTPRSLSAIDRAGAEVKSRTTLFRKIGDFRSLEKISREVKSITIIGGGF 310
Query 98 IGLE----LGSVWRNLGAEVTVVEFCD-----KIIPALDAEIGRAFQKLLERQGIKFMFG 148
+G E LG R LG EV + F + KI+P + + + R+G+K M
Sbjct 311 LGSELACALGRKARALGTEVIQL-FPEKGNMGKILPEY---LSNWTMEKVRREGVKVM-P 365
Query 149 TKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVGRRP 189
+V S G L +K D +V+ D ++ AVG P
Sbjct 366 NAIVQSVGVSSGKLL----IKLKDGRKVETDHIVAAVGLEP 402
> CE08028_2
Length=6284
Score = 31.6 bits (70), Expect = 1.4, Method: Composition-based stats.
Identities = 13/40 (32%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 140 RQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCD 179
++G + +V K DG +TL+ +NV DA E +C+
Sbjct 3504 KEGKEVEMSARVRAEHKDDGTLTLTFDNVTQADAGEYRCE 3543
> Hs22041912
Length=590
Score = 31.2 bits (69), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 32/111 (28%), Positives = 51/111 (45%), Gaps = 10/111 (9%)
Query 1 IVSALSQGIEGLFKRNKVDYLKGTGRLAGPHAV-----QVQPIDA-GNPQML--MAKNVI 52
+ S SQ LF+R+ + L+G+ RL+ + V V P ++ +P+++ + + VI
Sbjct 341 LASPHSQQAPFLFQRSHIYDLQGSKRLSTMYCVPKSTASVHPQNSFQHPEVVRGVVEKVI 400
Query 53 LATGSEPAPLAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELG 103
G+ PAP G L G L P P+HL V+G LG
Sbjct 401 GFAGAPPAPEHLGVLGAPPAP--EHLGVLGAPPAPEHLGVLGAPPAPEHLG 449
> SPAC3A11.07
Length=551
Score = 30.8 bits (68), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 42/151 (27%), Positives = 63/151 (41%), Gaps = 25/151 (16%)
Query 89 HLVVVGGGVIGLE------------LGSVWRNLGAE--VTVVEFCDKIIPALDAEIGRAF 134
H VVVGGG G+E L S + L + VT+VE ++P A++
Sbjct 256 HTVVVGGGPTGMEFAGEMADFIEDDLKSWYPELADDFAVTLVEALPSVLPMFSAKLRDYT 315
Query 135 QKLLERQGIKFMFGTKVVGSQKADGGVTLSLENVKSGDASEVQCDVVLVAVG---RRPYT 191
Q L + IK T + + + + ++N EV +LV G RP T
Sbjct 316 QSLFDSSHIKIRTNTAL--KKVTAENIHVEVKNPDGSKQEEVIPYGLLVWAGGNRARPLT 373
Query 192 KDL--GLEELGINLDNRGRVGVNEQMLVPNY 220
K L G EE +NR + V+E + + Y
Sbjct 374 KKLMEGSEE----QNNRRGLVVDEYLKLKGY 400
> 7291425
Length=1276
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/87 (28%), Positives = 36/87 (41%), Gaps = 3/87 (3%)
Query 62 LAGGALEVDEETIVSSTGALALPR---VPKHLVVVGGGVIGLELGSVWRNLGAEVTVVEF 118
+A G + V ET+ AL R H++ GG G S+ RNLG VV
Sbjct 435 VALGFIRVANETMCRPIRALTQSRGLDTANHVLSCFGGAGGQHACSIARNLGIAKVVVHK 494
Query 119 CDKIIPALDAEIGRAFQKLLERQGIKF 145
I+ A + Q+ E G++F
Sbjct 495 YAGILSAYGMALADVVQEFQEPNGLEF 521
> Hs22045508
Length=444
Score = 28.9 bits (63), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 17/45 (37%), Positives = 24/45 (53%), Gaps = 0/45 (0%)
Query 62 LAGGALEVDEETIVSSTGALALPRVPKHLVVVGGGVIGLELGSVW 106
L+ G+ + T VS +GA PRVP+ L V G L G++W
Sbjct 344 LSFGSWGIWRLTAVSRSGATGAPRVPRALGVEGSPGAWLLAGALW 388
> At3g14270
Length=1791
Score = 28.9 bits (63), Expect = 8.8, Method: Composition-based stats.
Identities = 18/68 (26%), Positives = 34/68 (50%), Gaps = 0/68 (0%)
Query 27 LAGPHAVQVQPIDAGNPQMLMAKNVILATGSEPAPLAGGALEVDEETIVSSTGALALPRV 86
L G + P +G+ ++L+ +N+I A + P + A + E + + T LAL V
Sbjct 1628 LKGSSRARYNPDSSGSNKVLLDQNLIEAMPTSPIFVGNKAKRLLERAVWNDTAFLALGDV 1687
Query 87 PKHLVVVG 94
+ ++VG
Sbjct 1688 MDYSLLVG 1695
Lambda K H
0.316 0.138 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4183546302
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40