bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0306_orf1
Length=117
Score E
Sequences producing significant alignments: (Bits) Value
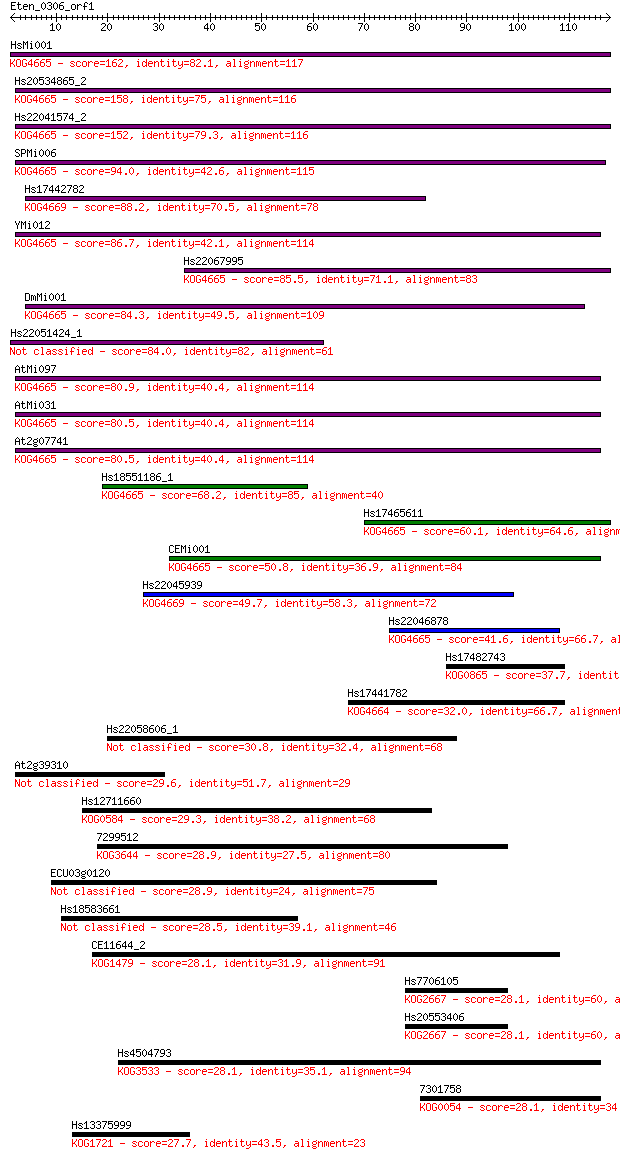
HsMi001 162 1e-40
Hs20534865_2 158 3e-39
Hs22041574_2 152 2e-37
SPMi006 94.0 6e-20
Hs17442782 88.2 3e-18
YMi012 86.7 1e-17
Hs22067995 85.5 2e-17
DmMi001 84.3 4e-17
Hs22051424_1 84.0 7e-17
AtMi097 80.9 6e-16
AtMi031 80.5 8e-16
At2g07741 80.5 8e-16
Hs18551186_1 68.2 4e-12
Hs17465611 60.1 1e-09
CEMi001 50.8 5e-07
Hs22045939 49.7 1e-06
Hs22046878 41.6 4e-04
Hs17482743 37.7 0.006
Hs17441782 32.0 0.30
Hs22058606_1 30.8 0.70
At2g39310 29.6 1.4
Hs12711660 29.3 2.1
7299512 28.9 2.3
ECU03g0120 28.9 2.4
Hs18583661 28.5 2.8
CE11644_2 28.1 4.1
Hs7706105 28.1 4.4
Hs20553406 28.1 4.4
Hs4504793 28.1 4.4
7301758 28.1 4.6
Hs13375999 27.7 5.8
> HsMi001
Length=226
Score = 162 bits (411), Expect = 1e-40, Method: Compositional matrix adjust.
Identities = 96/117 (82%), Positives = 106/117 (90%), Gaps = 0/117 (0%)
Query 1 AGAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHL 60
AGAVI GFR+K K +LAHFLPQGTPTPLIP+LVIIETISL IQP+ALAVRLTANITAGHL
Sbjct 110 AGAVIMGFRSKIKNALAHFLPQGTPTPLIPMLVIIETISLLIQPMALAVRLTANITAGHL 169
Query 61 LIHLIGGATLALISISTTTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHDNT 117
L+HLIG ATLA+ +I+ + LI FTILILLTILE AVA+IQAYVFTLLVSLYLHDNT
Sbjct 170 LMHLIGSATLAMSTINLPSTLIIFTILILLTILEIAVALIQAYVFTLLVSLYLHDNT 226
> Hs20534865_2
Length=122
Score = 158 bits (399), Expect = 3e-39, Method: Compositional matrix adjust.
Identities = 87/116 (75%), Positives = 95/116 (81%), Gaps = 0/116 (0%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
G VITGFR KTK SLAHFLPQGT PLIPI V ETISLFIQP ALAVRLTAN+ A HLL
Sbjct 7 GTVITGFRFKTKNSLAHFLPQGTRIPLIPIPVFTETISLFIQPTALAVRLTANLIASHLL 66
Query 62 IHLIGGATLALISISTTTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHDNT 117
+ LIGGATL L + + TA ITF ILILLT+L+ AVA+IQAYVFTLLVSLY +DNT
Sbjct 67 MRLIGGATLVLSTTNLPTASITFIILILLTMLQSAVALIQAYVFTLLVSLYFYDNT 122
> Hs22041574_2
Length=134
Score = 152 bits (383), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 92/116 (79%), Positives = 99/116 (85%), Gaps = 0/116 (0%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
G VITGF KTK SLAHFLPQGTP PLIPILVI ETISLFIQP+ LAV+LTA ITAGHLL
Sbjct 19 GTVITGFCFKTKVSLAHFLPQGTPMPLIPILVITETISLFIQPMVLAVQLTAKITAGHLL 78
Query 62 IHLIGGATLALISISTTTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHDNT 117
IHLI GATLAL +IS TA I ILILLTIL+FA+A+IQAYVFTLLV LYL+DNT
Sbjct 79 IHLIRGATLALSTISLPTASIALIILILLTILKFAIALIQAYVFTLLVGLYLYDNT 134
> SPMi006
Length=257
Score = 94.0 bits (232), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 49/118 (41%), Positives = 75/118 (63%), Gaps = 3/118 (2%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
GA I G + FLP GTPTPLIP+LV+IE +S + ++L +RL ANI AGHL
Sbjct 136 GATILGLQQHKAKVFGLFLPSGTPTPLIPLLVLIEFVSYIARGLSLGIRLGANIIAGHLT 195
Query 62 IHLIGGATLALISISTTTALITF---TILILLTILEFAVAIIQAYVFTLLVSLYLHDN 116
+ ++GG + ++ T +I F T+L+ +++LEF +A IQAYVF +L +++D+
Sbjct 196 MSILGGLIFTFMGLNLITFIIGFLPITVLVAISLLEFGIAFIQAYVFAILTCGFINDS 253
> Hs17442782
Length=101
Score = 88.2 bits (217), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 55/81 (67%), Positives = 57/81 (70%), Gaps = 7/81 (8%)
Query 4 VITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLLIH 63
VITGFR KTK SLAHFLPQGT PLIPI V ETISLFIQP ALAVRLTANITA I
Sbjct 14 VITGFRFKTKNSLAHFLPQGTRIPLIPIPVFTETISLFIQPTALAVRLTANITA----IF 69
Query 64 LIGGATLA---LISISTTTAL 81
G A + L+SIS T L
Sbjct 70 AAGKAAVGLALLVSISNTYDL 90
> YMi012
Length=259
Score = 86.7 bits (213), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 48/117 (41%), Positives = 72/117 (61%), Gaps = 3/117 (2%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
G I G + F+P GTP PL+P+LVIIET+S F + I+L +RL +NI AGHLL
Sbjct 138 GNTILGLYKHGWVFFSLFVPAGTPLPLVPLLVIIETLSYFARAISLGLRLGSNILAGHLL 197
Query 62 IHLIGGATLALISISTTTALITFT---ILILLTILEFAVAIIQAYVFTLLVSLYLHD 115
+ ++ G T + I+ T + F +++ + +LEFA+ IIQ YV+ +L + YL D
Sbjct 198 MVILAGLTFNFMLINLFTLVFGFVPLAMILAIMMLEFAIGIIQGYVWAILTASYLKD 254
> Hs22067995
Length=128
Score = 85.5 bits (210), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 59/83 (71%), Positives = 66/83 (79%), Gaps = 4/83 (4%)
Query 35 IETISLFIQPIALAVRLTANITAGHLLIHLIGGATLALISISTTTALITFTILILLTILE 94
I SLFIQPIALAVRLTA ITAGHLLIHLIG ATL L +IS TA I+ ILILL+ILE
Sbjct 31 INKSSLFIQPIALAVRLTATITAGHLLIHLIGDATLVLSTISIPTASISLIILILLSILE 90
Query 95 FAVAIIQAYVFTLLVSLYLHDNT 117
F +A+IQAYVFTLL L+DN+
Sbjct 91 FVIALIQAYVFTLL----LNDNS 109
> DmMi001
Length=224
Score = 84.3 bits (207), Expect = 4e-17, Method: Compositional matrix adjust.
Identities = 54/111 (48%), Positives = 74/111 (66%), Gaps = 7/111 (6%)
Query 4 VITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLLIH 63
++ G+ N T+ AH +PQGTP L+P +V IETIS I+P LAVRLTAN+ AGHLL+
Sbjct 114 MLYGWINHTQHMFAHLVPQGTPAILMPFMVCIETISNIIRPGTLAVRLTANMIAGHLLLT 173
Query 64 LIGGATLALISISTTTALITFTIL--ILLTILEFAVAIIQAYVFTLLVSLY 112
L+G S + L+TF ++ I L +LE AVA+IQ+YVF +L +LY
Sbjct 174 LLGNT-----GSSMSYMLMTFLLMAQIALLVLESAVAMIQSYVFAVLSTLY 219
> Hs22051424_1
Length=183
Score = 84.0 bits (206), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 50/61 (81%), Positives = 54/61 (88%), Gaps = 0/61 (0%)
Query 1 AGAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHL 60
G VITG R+KTK +LAH LPQGTP PLIPIL+IIETISL IQP+ALAVRLTANITAGHL
Sbjct 123 GGTVITGLRSKTKNALAHLLPQGTPAPLIPILIIIETISLLIQPVALAVRLTANITAGHL 182
Query 61 L 61
L
Sbjct 183 L 183
> AtMi097
Length=349
Score = 80.9 bits (198), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 69/118 (58%), Gaps = 4/118 (3%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
G I GF+ + LP G P PL P LV++E IS + ++L +RL AN+ AGH L
Sbjct 227 GITIVGFQRHGLHFFSFLLPAGVPLPLAPFLVLLELISYCFRALSLGIRLFANMMAGHSL 286
Query 62 IHLIGGATLALISIST----TTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHD 115
+ ++ G ++ ++ AL I++ LT LE VAI+QAYVFT+L+ +YL+D
Sbjct 287 VKILSGFAWTMLCMNDIFYFIGALGPLFIVLALTGLELGVAILQAYVFTILICIYLND 344
> AtMi031
Length=385
Score = 80.5 bits (197), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 69/118 (58%), Gaps = 4/118 (3%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
G I GF+ + LP G P PL P LV++E IS + ++L +RL AN+ AGH L
Sbjct 263 GITIVGFQRHGLHFFSFLLPAGVPLPLAPFLVLLELISYCFRALSLGIRLFANMMAGHSL 322
Query 62 IHLIGGATLALISIST----TTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHD 115
+ ++ G ++ ++ AL I++ LT LE VAI+QAYVFT+L+ +YL+D
Sbjct 323 VKILSGFAWTMLCMNDIFYFIGALGPLFIVLALTGLELGVAILQAYVFTILICIYLND 380
> At2g07741
Length=385
Score = 80.5 bits (197), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 46/118 (38%), Positives = 69/118 (58%), Gaps = 4/118 (3%)
Query 2 GAVITGFRNKTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLL 61
G I GF+ + LP G P PL P LV++E IS + ++L +RL AN+ AGH L
Sbjct 263 GITIVGFQRHGLHFFSFLLPAGVPLPLAPFLVLLELISYCFRALSLGIRLFANMMAGHSL 322
Query 62 IHLIGGATLALISIST----TTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHD 115
+ ++ G ++ ++ AL I++ LT LE VAI+QAYVFT+L+ +YL+D
Sbjct 323 VKILSGFAWTMLCMNDIFYFIGALGPLFIVLALTGLELGVAILQAYVFTILICIYLND 380
> Hs18551186_1
Length=58
Score = 68.2 bits (165), Expect = 4e-12, Method: Compositional matrix adjust.
Identities = 34/40 (85%), Positives = 37/40 (92%), Gaps = 0/40 (0%)
Query 19 FLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAG 58
FL QGTP PLIPILV+IETI LFIQP+ALA+RLTANITAG
Sbjct 19 FLSQGTPMPLIPILVVIETIHLFIQPVALAMRLTANITAG 58
> Hs17465611
Length=103
Score = 60.1 bits (144), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 31/48 (64%), Positives = 35/48 (72%), Gaps = 0/48 (0%)
Query 70 LALISISTTTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHDNT 117
L LI + TF IL+LLT LEFAVA+ QAY+FTLLVSLY HDNT
Sbjct 56 LTLIEYGCNFSSSTFIILVLLTTLEFAVALFQAYIFTLLVSLYFHDNT 103
> CEMi001
Length=199
Score = 50.8 bits (120), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 31/84 (36%), Positives = 52/84 (61%), Gaps = 6/84 (7%)
Query 32 LVIIETISLFIQPIALAVRLTANITAGHLLIHLIGGATLALISISTTTALITFTILILLT 91
+++IE +S F +P+AL VRLT NIT GHL+ ++ + +S I +IL ++
Sbjct 122 MLLIEIVSEFSRPLALTVRLTVNITVGHLVSMMLYQG----LELSMGDQYIWLSILAIM- 176
Query 92 ILEFAVAIIQAYVFTLLVSLYLHD 115
+E V IQ+Y+F+ L+ LYL++
Sbjct 177 -MECFVFFIQSYIFSRLIFLYLNE 199
> Hs22045939
Length=164
Score = 49.7 bits (117), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 42/85 (49%), Positives = 51/85 (60%), Gaps = 13/85 (15%)
Query 27 PLIPILVIIETISLFIQPIALAVRLTANITAGH----------LLIHLIGGATLALISIS 76
PL PILVII+TIS IQ +ALAV LTANITA H L +H I G L+L I+
Sbjct 51 PLTPILVIIQTISPLIQLMALAVHLTANITADHSQIRCGPYPNLCLHAIKGIILSLFIIN 110
Query 77 TTTAL-ITFTI--LILLTILEFAVA 98
T L I FT+ +I +T+L FA
Sbjct 111 TLIPLNIHFTLAFIIPITLLVFAAC 135
> Hs22046878
Length=288
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 22/33 (66%), Positives = 25/33 (75%), Gaps = 0/33 (0%)
Query 75 ISTTTALITFTILILLTILEFAVAIIQAYVFTL 107
+ + A IT I ILLT LEFAVA+IQAYVFTL
Sbjct 1 MESKPASITLIIPILLTTLEFAVALIQAYVFTL 33
> Hs17482743
Length=194
Score = 37.7 bits (86), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 18/23 (78%), Positives = 21/23 (91%), Gaps = 0/23 (0%)
Query 86 ILILLTILEFAVAIIQAYVFTLL 108
ILI+LTI EF +A+IQAYVFTLL
Sbjct 128 ILIILTIFEFVIALIQAYVFTLL 150
> Hs17441782
Length=102
Score = 32.0 bits (71), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 28/42 (66%), Positives = 31/42 (73%), Gaps = 0/42 (0%)
Query 67 GATLALISISTTTALITFTILILLTILEFAVAIIQAYVFTLL 108
G TL L +IS A I F ILILLTILEF +A+IQAYV TLL
Sbjct 9 GPTLVLSTISLPIASIAFIILILLTILEFTIALIQAYVLTLL 50
> Hs22058606_1
Length=634
Score = 30.8 bits (68), Expect = 0.70, Method: Composition-based stats.
Identities = 22/69 (31%), Positives = 34/69 (49%), Gaps = 3/69 (4%)
Query 20 LPQGTPTPLIPILVIIETISLFIQ-PIALAVRLTANITAGHLLIHLIGGATLALISISTT 78
LP+ PT L+P L ++ I F+Q P R+ + + G +I L L L S+ST
Sbjct 60 LPRAMPTILVPKLKALKMIPPFVQLPSCFVSRIISGVILG--VIALRPSKGLVLDSLSTY 117
Query 79 TALITFTIL 87
+ T +L
Sbjct 118 LKIHTSVVL 126
> At2g39310
Length=458
Score = 29.6 bits (65), Expect = 1.4, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 4/33 (12%)
Query 2 GAVITGFRNKTKASL----AHFLPQGTPTPLIP 30
G + GF K A++ A+F P TPTPLIP
Sbjct 278 GHRLVGFHGKEDAAIDALGAYFGPVPTPTPLIP 310
> Hs12711660
Length=2382
Score = 29.3 bits (64), Expect = 2.1, Method: Composition-based stats.
Identities = 26/72 (36%), Positives = 34/72 (47%), Gaps = 10/72 (13%)
Query 15 SLAHFLPQGT--PTPLIPILVIIETISLFIQPIALAVRLTANITAGHLLIHLIGGATLAL 72
S AHFLP G PTPL+P + + PI+ TA L I + G T L
Sbjct 820 SGAHFLPVGQPLPTPLLPQYPVSQI------PISTPHVSTAQTGFSSLPITMAAGITQPL 873
Query 73 ISI--STTTALI 82
+++ S TTA I
Sbjct 874 LTLASSATTAAI 885
> 7299512
Length=387
Score = 28.9 bits (63), Expect = 2.3, Method: Composition-based stats.
Identities = 22/84 (26%), Positives = 42/84 (50%), Gaps = 7/84 (8%)
Query 18 HFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLLIHLIGGATLALISIST 77
H P+ LI V++ IS +I+P A+ R+T +T+ L + +L +S
Sbjct 266 HLFHTYIPSALI---VVMSWISFWIKPEAIPARVTLGVTSLLTLATQNTQSQQSLPPVSY 322
Query 78 TTALITF----TILILLTILEFAV 97
A+ + ++ + L+++EFAV
Sbjct 323 VKAIDVWMSSCSVFVFLSLMEFAV 346
> ECU03g0120
Length=276
Score = 28.9 bits (63), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/77 (23%), Positives = 40/77 (51%), Gaps = 13/77 (16%)
Query 9 RNKTKASLAHFLPQGTPTPLIPILVIIETI--SLFIQPIALAVRLTANITAGHLLIHLIG 66
+N+ +AS++ +G P +I ++++ TI + + PI+ + G+ L+H++G
Sbjct 29 KNEKEASVS----EGKPAAVIDSILLLSTILVAWILHPISKKI-------FGNFLLHIVG 77
Query 67 GATLALISISTTTALIT 83
L + ST + L +
Sbjct 78 WGNLGYLVFSTISNLFS 94
> Hs18583661
Length=455
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Query 11 KTKASLAHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANIT 56
+ AS+AHF P P PL+ L E SL + + LA LT+ T
Sbjct 242 RGSASMAHFFPISIPEPLLGELA-EEAASLRDRALVLASSLTSRRT 286
> CE11644_2
Length=737
Score = 28.1 bits (61), Expect = 4.1, Method: Composition-based stats.
Identities = 29/93 (31%), Positives = 47/93 (50%), Gaps = 6/93 (6%)
Query 17 AHFLPQGTPTPLIPILVIIETISLFIQPIALAVRLTANITAGHLLIHLIGGATLALISIS 76
++F T + IP LV I++FI A+ LT + +++ L+ T+ I I
Sbjct 374 SNFQNSMTISAQIPSLVF-SVINIFI---AVKGDLTRGMKICLIVVQLMVIVTVVFIYID 429
Query 77 TTTALITFTILILLTI--LEFAVAIIQAYVFTL 107
T+T + TF++L L TI L A + Q +F L
Sbjct 430 TSTWIATFSMLTLGTIVVLNAANGLFQNSMFGL 462
> Hs7706105
Length=378
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 78 TTALITFTILILLTILEFAV 97
T +LI FT + LLTI+EF+V
Sbjct 35 TVSLIAFTTMALLTIMEFSV 54
> Hs20553406
Length=377
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 12/20 (60%), Positives = 16/20 (80%), Gaps = 0/20 (0%)
Query 78 TTALITFTILILLTILEFAV 97
T +LI FT + LLTI+EF+V
Sbjct 35 TVSLIAFTTMALLTIMEFSV 54
> Hs4504793
Length=2701
Score = 28.1 bits (61), Expect = 4.4, Method: Composition-based stats.
Identities = 33/110 (30%), Positives = 50/110 (45%), Gaps = 17/110 (15%)
Query 22 QGTPTPLIPIL----VIIETISLF-------IQP--IALAVRLTANITAGHLLIHLIGGA 68
+GT +PL +L V I T LF I+P +++ +R I G LI L+G A
Sbjct 2256 EGTLSPLFSVLLWIAVAICTSMLFFFSKPVGIRPFLVSIMLRSIYTIGLGPTLI-LLGAA 2314
Query 69 TLA---LISISTTTALITFTILILLTILEFAVAIIQAYVFTLLVSLYLHD 115
L + +S TFT IL+ A AYV ++ L++H+
Sbjct 2315 NLCNKIVFLVSFVGNRGTFTRGYRAVILDMAFLYHVAYVLVCMLGLFVHE 2364
> 7301758
Length=1302
Score = 28.1 bits (61), Expect = 4.6, Method: Composition-based stats.
Identities = 12/35 (34%), Positives = 22/35 (62%), Gaps = 0/35 (0%)
Query 81 LITFTILILLTILEFAVAIIQAYVFTLLVSLYLHD 115
+I +T LIL T++ + + ++ TL +SL +HD
Sbjct 764 MILYTFLILCTLIFYVLRTFGFFMMTLRISLRIHD 798
> Hs13375999
Length=498
Score = 27.7 bits (60), Expect = 5.8, Method: Composition-based stats.
Identities = 10/23 (43%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 13 KASLAHFLPQGTPTPLIPILVII 35
K H +P GTPTPL P++ ++
Sbjct 468 KHERTHPVPMGTPTPLEPLVALL 490
Lambda K H
0.329 0.142 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1171470346
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40