bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0282_orf1
Length=233
Score E
Sequences producing significant alignments: (Bits) Value
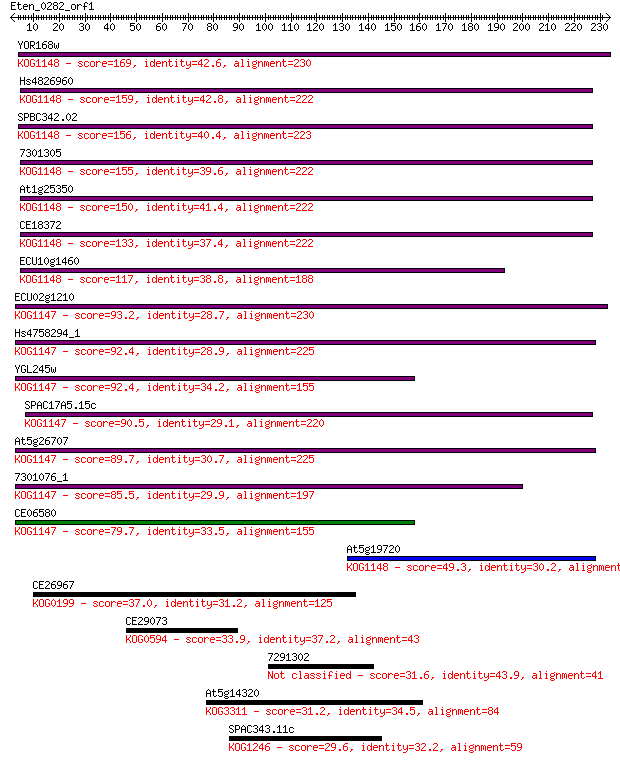
YOR168w 169 3e-42
Hs4826960 159 6e-39
SPBC342.02 156 4e-38
7301305 155 5e-38
At1g25350 150 2e-36
CE18372 133 4e-31
ECU10g1460 117 1e-26
ECU02g1210 93.2 4e-19
Hs4758294_1 92.4 7e-19
YGL245w 92.4 7e-19
SPAC17A5.15c 90.5 3e-18
At5g26707 89.7 4e-18
7301076_1 85.5 9e-17
CE06580 79.7 4e-15
At5g19720 49.3 7e-06
CE26967 37.0 0.031
CE29073 33.9 0.29
7291302 31.6 1.3
At5g14320 31.2 1.6
SPAC343.11c 29.6 5.5
> YOR168w
Length=809
Score = 169 bits (429), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 98/236 (41%), Positives = 135/236 (57%), Gaps = 9/236 (3%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R+ Y+W + + + R Q E+ RLN+T TV+SKRK+ LV+EK V GWDDPRL TL A+
Sbjct 465 SRESYEWLCDQVHVFRPAQREYGRLNITGTVLSKRKIAQLVDEKFVRGWDDPRLFTLEAI 524
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRGVPP AI F +GV + IQ+ E +R+YL++ PR V P+ V + N
Sbjct 525 RRRGVPPGAILSFINTLGVTTSTTNIQVVRFESAVRKYLEDTTPRLMFVLDPVEVVVDNL 584
Query 124 G--AEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMEN-PPKEFHRLSPGKEVRL-RS 179
EE+ TIP P P G R + FTN+ YI+ DF EN KEF RL+P + V L +
Sbjct 585 SDDYEELATIPYRPGTPEFGERTVPFTNKFYIERSDFSENVDDKEFFRLTPNQPVGLIKV 644
Query 180 AYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP--AKYAVP 233
++ + +EKD G I I +YD + S +K K I W+P +KY P
Sbjct 645 SHTVSFKSLEKDEAGKIIRIHVNYDNKVEEGSKP---KKPKTYIQWVPISSKYNSP 697
> Hs4826960
Length=775
Score = 159 bits (401), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 95/225 (42%), Positives = 126/225 (56%), Gaps = 10/225 (4%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W AL + Q E+ RLN+ Y V+SKRK+L LV V WDDPRL TL+ALR
Sbjct 464 RSSYFWLCNALDVYCPVQWEYGRLNLHYAVVSKRKILQLVATGAVRDWDDPRLFTLTALR 523
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFG 124
RRG PP AI +FC RVGV +T++ LLE C+R+ L++ APR AV + L V ITNF
Sbjct 524 RRGFPPEAINNFCARVGVTVAQTTMEPHLLEACVRDVLNDTAPRAMAVLESLRVIITNFP 583
Query 125 AEEVLTI--PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR-SAY 181
A + L I P+ P G Q+ F ++I+ DF E P F RL+ G+ V LR + Y
Sbjct 584 AAKSLDIQVPNFPADETKGFHQVPFAPIVFIERTDFKEEPEPGFKRLAWGQPVGLRHTGY 643
Query 182 WIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
I+ V K +G + ++ + C A G K K IHW+
Sbjct 644 VIELQHVVKGPSGCVESL-------EVTCRRADAGEKPKAFIHWV 681
> SPBC342.02
Length=811
Score = 156 bits (394), Expect = 4e-38, Method: Compositional matrix adjust.
Identities = 90/226 (39%), Positives = 122/226 (53%), Gaps = 11/226 (4%)
Query 4 NRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSAL 63
+R Y+W AL + Q E+ RLNV T+MSKRK++ LV E V GW+DPRL TL AL
Sbjct 472 SRVSYEWLCNALEVYCPAQREYGRLNVVGTLMSKRKIMKLVKEGYVHGWNDPRLYTLVAL 531
Query 64 RRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRRGVPP AI +F VGV S I++ E C+R++L+ PR + P+ V + N
Sbjct 532 RRRGVPPGAILEFVSEVGVTTAVSNIEVARFENCVRKFLENSVPRLMFLPDPIKVTLENL 591
Query 124 --GAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRL-RSA 180
E + IP +PK P+ GSR T +YID DF E +F RL+ G+ V L R++
Sbjct 592 DDSYREQIEIPFNPKDPSMGSRSAFLTKHIYIDRSDFREEASSDFFRLTLGQPVGLFRAS 651
Query 181 YWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
+ + V K+ G II YD A +K K I W+
Sbjct 652 HPVVAKRVVKNDEGEPIEIIAEYD--------ASSSKKPKTFIQWV 689
> 7301305
Length=759
Score = 155 bits (393), Expect = 5e-38, Method: Compositional matrix adjust.
Identities = 88/226 (38%), Positives = 123/226 (54%), Gaps = 11/226 (4%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W ALGI Q E+ RLN+ Y ++SKRK+ L+ E++V WDDPRL TL+ALR
Sbjct 448 RSSYYWLCNALGIYCPVQWEYGRLNMNYALVSKRKIAKLITEQIVHDWDDPRLFTLTALR 507
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF- 123
RRG P AI +FC ++GV + +LE +R+ L+ APRR V +PL V I NF
Sbjct 508 RRGFPAEAINNFCAQMGVTGAQIAVDPAMLEAAVRDVLNVTAPRRLVVLEPLKVTIKNFP 567
Query 124 -GAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSA-Y 181
A L +P P+ P G+ ++ +YI+ DF P K + RL+P + V LR A
Sbjct 568 HAAPVQLEVPDFPQNPQQGTHKITLDKVIYIEQGDFKLEPEKGYRRLAPKQSVGLRHAGL 627
Query 182 WIKCNEVEKD-ANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
I +E+ KD A G + +IC+ P A K K + W+
Sbjct 628 VISVDEIVKDPATGQVVELICTSQP-------AEQAEKPKAFVQWV 666
> At1g25350
Length=786
Score = 150 bits (379), Expect = 2e-36, Method: Compositional matrix adjust.
Identities = 92/229 (40%), Positives = 123/229 (53%), Gaps = 15/229 (6%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W +L + E++RLNVT TVMSKRKL +V K V GWDDPRL TLS LR
Sbjct 467 RASYYWLLHSLSLYMPYVWEYSRLNVTNTVMSKRKLNYIVTNKYVDGWDDPRLLTLSGLR 526
Query 65 RRGVPPAAIRDFCERVGVARRP-STIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF 123
RRGV AI F +G+ R S I + LE IRE L++ APR V PL V ITN
Sbjct 527 RRGVTSTAINAFVRGIGITRSDGSMIHVSRLEHHIREELNKTAPRTMVVLNPLKVVITNL 586
Query 124 GAEEVLTI-----PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR 178
+++++ + P + ++ F+ +YID DF K+++ L+PGK V LR
Sbjct 587 ESDKLIELDAKRWPDAQNDDPSAFYKVPFSRVVYIDQSDFRMKDSKDYYGLAPGKSVLLR 646
Query 179 SAYWIKC-NEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
A+ IKC N V D N + I YDP+ + K KG +HW+
Sbjct 647 YAFPIKCTNVVFADDNETVREIHAEYDPEKKS--------KPKGVLHWV 687
> CE18372
Length=786
Score = 133 bits (334), Expect = 4e-31, Method: Compositional matrix adjust.
Identities = 83/230 (36%), Positives = 123/230 (53%), Gaps = 17/230 (7%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
R Y W AL I Q E+ RLNV YTV+SKRK+L L+ K V WDDPRL TL+ALR
Sbjct 470 RSSYYWLCNALDIYCPVQWEYGRLNVNYTVVSKRKILKLITTKTVNDWDDPRLFTLTALR 529
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF- 123
RRG+P AI F ++G+ I +L+ +R+YL+ APR AV + L + I NF
Sbjct 530 RRGIPSEAINRFVAKLGLTMSQMVIDPHVLDATVRDYLNIHAPRTMAVLEGLKLTIENFS 589
Query 124 -----GAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFM-ENPPKEFHRLSPGKEVRL 177
+ +V PSDP P S ++ E++I+ D+ ++ K F RL+P + V L
Sbjct 590 ELNLPSSVDVPDFPSDPTDPRKHS--VSVDREIFIEKSDYKPDDSDKSFRRLTPKQAVGL 647
Query 178 RS-AYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
+ ++ + KDA G++T ++ + + + K K IHW+
Sbjct 648 KHIGLVLRFVKEVKDAEGHVTEVVVKAEKLS-------EKDKPKAFIHWV 690
> ECU10g1460
Length=697
Score = 117 bits (294), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 73/188 (38%), Positives = 106/188 (56%), Gaps = 5/188 (2%)
Query 5 RQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR 64
++ Y+W L I + Q EF+RLN++ TV+SKRKLL L +K DDPRL T+ +R
Sbjct 412 QESYNWLLVQLEIYKPIQWEFSRLNISNTVLSKRKLLPL--KKYGIELDDPRLFTIKGMR 469
Query 65 RRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFG 124
RRG PP AI FC +G +T+ ++ LE +R+ L+ + R VK+PL V I N
Sbjct 470 RRGFPPEAINQFCRSLGFTFAETTVDVKKLENFVRDNLNRTSRRIMCVKEPLKVTIMN-S 528
Query 125 AEEVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAYWIK 184
++IP P ++ R + FT +YI+ DFME K+F RL+P + V L Y I+
Sbjct 529 TPCSISIPDLPG--SSVVRDVPFTPVIYIEKSDFMEKGDKDFLRLTPEQPVGLYMLYPIR 586
Query 185 CNEVEKDA 192
+V D
Sbjct 587 VVKVTPDG 594
> ECU02g1210
Length=642
Score = 93.2 bits (230), Expect = 4e-19, Method: Compositional matrix adjust.
Identities = 66/235 (28%), Positives = 124/235 (52%), Gaps = 25/235 (10%)
Query 3 RNRQLYDWFQEALGI-TRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLS 61
RN+Q Y WF + L + R + +F+RLN TV+SKRKL V+ V+GWDDPRL+T++
Sbjct 351 RNQQYY-WFIDNLRLRNRPKIHDFSRLNFENTVLSKRKLKYYVDNGFVSGWDDPRLATIA 409
Query 62 ALRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKIT 121
++R G+ A+R++ GV+++ TI + + R+ +D + R F V+Q V+++
Sbjct 410 GIKRLGMNMEALREYILMQGVSQKTCTISWDKVWAINRKKIDPVSARYFCVQQRDAVEVS 469
Query 122 -NFGAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAED---FMENPPKEFHRLSPGKEVRL 177
+ +E + +P K G++++ +++++ + ED +N +EF ++ G +
Sbjct 470 IDNTSEYTMDVPKHKKNGDLGTKEVFYSSQILLSQEDGRVLQDN--EEFTLMNWGNAI-- 525
Query 178 RSAYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLPAKYAV 232
+K VE NG +T + S +P D + K + W+ + +V
Sbjct 526 -----VKSKTVE---NGTVTKMEVSLNPDG-------DFKLTKNKMSWVSKRGSV 565
> Hs4758294_1
Length=941
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 65/229 (28%), Positives = 113/229 (49%), Gaps = 16/229 (6%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y W EALGI + E++RLN+ TV+SKRKL VNE +V GWDDPR T+
Sbjct 330 RDEQFY-WIIEALGIRKPYIWEYSRLNLNNTVLSKRKLTWFVNEGLVDGWDDPRFPTVRG 388
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ + ++ +
Sbjct 389 VLRRGMTVEGLKQFIAAQGSSRSVVNMEWDKIWAFNKKVIDPVAPRYVALLKKEVIPVNV 448
Query 123 FGA-EEVLTIPSDPKRPAAGSRQLAFTNELYI---DAEDFMENPPKEFHRLSPGKEVRLR 178
A EE+ + PK P G + + ++ +++I DAE F E G+ V
Sbjct 449 PEAQEEMKEVAKHPKNPEVGLKPVWYSPKVFIEGADAETFSE-----------GEMVTFI 497
Query 179 SAYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
+ + ++ K+A+G I ++ ++ + + + T H LP
Sbjct 498 NWGNLNITKIHKNADGKIISLDAKFNLENKDYKKTTKVTWLAETTHALP 546
> YGL245w
Length=724
Score = 92.4 bits (228), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 53/160 (33%), Positives = 88/160 (55%), Gaps = 8/160 (5%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
RN Q YDW +AL + + +FAR+N T++SKRKL +V++ +V WDDPR T+
Sbjct 423 RNAQ-YDWMLQALRLRKVHIWDFARINFVRTLLSKRKLQWMVDKDLVGNWDDPRFPTVRG 481
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+RRRG+ +R+F G +R ++ L+ ++ +D APR A+ P VKI
Sbjct 482 VRRRGMTVEGLRNFVLSQGPSRNVINLEWNLIWAFNKKVIDPIAPRHTAIVNP--VKIHL 539
Query 123 FGAE-----EVLTIPSDPKRPAAGSRQLAFTNELYIDAED 157
G+E ++ P K PA G +++ + ++ +D +D
Sbjct 540 EGSEAPQEPKIEMKPKHKKNPAVGEKKVIYYKDIVVDKDD 579
> SPAC17A5.15c
Length=716
Score = 90.5 bits (223), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 64/226 (28%), Positives = 107/226 (47%), Gaps = 24/226 (10%)
Query 7 LYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALRRR 66
LY W +A+ + + EF+R+N T++SKRKL +V+ +V GWDDPR T+ +RRR
Sbjct 414 LYQWMIKAMNLRKIHVWEFSRMNFVRTLLSKRKLTEIVDHGLVWGWDDPRFPTVRGVRRR 473
Query 67 GVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNF--- 123
G+ A++ + G ++ T+ ++ +D APR AV+ +VK T
Sbjct 474 GMTIEALQQYIVSQGPSKNILTLDWTSFWATNKKIIDPVAPRHTAVESGDVVKATIVNGP 533
Query 124 GAEEVLTIPSDPKRPAAGSRQLAFTNELYI---DAEDFMENPPKEFHRLSPGKEVRLRSA 180
A P K P G+++ F NE+ I DA+ F ++ +EV L
Sbjct 534 AAPYAEDRPRHKKNPELGNKKSIFANEILIEQADAQSFKQD-----------EEVTLMDW 582
Query 181 YWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWL 226
E+ +DA+G +T++ L + D +K + + WL
Sbjct 583 GNAYVREINRDASGKVTSL-------KLELHLDGDFKKTEKKVTWL 621
> At5g26707
Length=719
Score = 89.7 bits (221), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 69/229 (30%), Positives = 112/229 (48%), Gaps = 21/229 (9%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
RN Q + ++ +G+ + + EF+RLN+ +T++SKRKLL V +V GWDDPR T+
Sbjct 418 RNAQYFKVLED-MGLRQVQLYEFSRLNLVFTLLSKRKLLWFVQTGLVDGWDDPRFPTVQG 476
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAV--KQPLLVKI 120
+ RRG+ A+ F G ++ + ++ + L + +D PR AV ++ +L +
Sbjct 477 IVRRGLKIEALIQFILEQGASKNLNLMEWDKLWSINKRIIDPVCPRHTAVVAERRVLFTL 536
Query 121 TNFGAEE--VLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLR 178
T+ G +E V IP K AG + FT ++++ D +S G+EV L
Sbjct 537 TD-GPDEPFVRMIPKHKKFEGAGEKATTFTKSIWLEEAD--------ASAISVGEEVTLM 587
Query 179 SAYWIKCNEVEKDANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
E+ KD G +TA+ LN + K+K T WLP
Sbjct 588 DWGNAIVKEITKDEEGRVTAL-----SGVLNLQGSVKTTKLKLT--WLP 629
> 7301076_1
Length=1128
Score = 85.5 bits (210), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 59/198 (29%), Positives = 100/198 (50%), Gaps = 10/198 (5%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y WF +AL + + ++RLN+T TV+SKRKL V+ +V GWDDPR T+
Sbjct 408 RDDQFY-WFIDALKLRKPYIWSYSRLNMTNTVLSKRKLTWFVDSGLVDGWDDPRFPTVRG 466
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITN 122
+ RRG+ +++F G ++ + + + ++ +D APR A+++ V +
Sbjct 467 IIRRGMTVEGLKEFIIAQGSSKSVVFMNWDKIWAFNKKVIDPIAPRYTALEKEKRVIVNV 526
Query 123 FGAE-EVLTIPSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAY 181
GA+ E + + PK + G + + +YID D L G+ +
Sbjct 527 AGAKVERIQVSVHPKDESLGKKTVLLGPRIYIDYVD--------AEALKEGENATFINWG 578
Query 182 WIKCNEVEKDANGNITAI 199
I +V KDA+GNIT++
Sbjct 579 NILIRKVNKDASGNITSV 596
> CE06580
Length=1149
Score = 79.7 bits (195), Expect = 4e-15, Method: Composition-based stats.
Identities = 52/158 (32%), Positives = 87/158 (55%), Gaps = 4/158 (2%)
Query 3 RNRQLYDWFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSA 62
R+ Q Y + +ALG+ R E+ARLN+T TVMSKRKL V+E V GWDDPRL T+
Sbjct 398 RDDQYY-FICDALGLRRPHIWEYARLNMTNTVMSKRKLTWFVDEGHVEGWDDPRLPTVRG 456
Query 63 LRRRGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAV--KQPLL-VK 119
+ RRG+ ++ F G +R ++ + + ++ +D APR A+ PL+ ++
Sbjct 457 VMRRGLTVEGLKQFIVAQGGSRSVVMMEWDKIWAFNKKVIDPVAPRYTALDSTSPLVSIE 516
Query 120 ITNFGAEEVLTIPSDPKRPAAGSRQLAFTNELYIDAED 157
+T+ +++ + PK GS+ + +L ++ D
Sbjct 517 LTDSISDDTSNVSLHPKNAEIGSKDVHKGKKLLLEQVD 554
> At5g19720
Length=170
Score = 49.3 bits (116), Expect = 7e-06, Method: Compositional matrix adjust.
Identities = 29/97 (29%), Positives = 44/97 (45%), Gaps = 9/97 (9%)
Query 132 PSDPKRPAAGSRQLAFTNELYIDAEDFMENPPKEFHRLSPGKEVRLRSAYWIKC-NEVEK 190
P P + ++ + +Y++ DF K ++ L+PGK V LR + IKC N V
Sbjct 9 PDAPNDDPSAFYKVPLSRVIYMEQSDFQNEDSKNYYGLAPGKSVLLRYTFPIKCTNVVFA 68
Query 191 DANGNITAIICSYDPQTLNCSVAPDGRKVKGTIHWLP 227
D + I YDP+ K KG +HW+P
Sbjct 69 DDTKTVCEIYVEYDPEK--------KIKPKGVLHWVP 97
> CE26967
Length=1043
Score = 37.0 bits (84), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 39/129 (30%), Positives = 53/129 (41%), Gaps = 21/129 (16%)
Query 10 WFQEALGITRTRQIEFARLNVTYTVMSKRKLLALVNEKVVAGWDDPRLSTLSALR----R 65
+ QEA +TR R RL Y V+ K + LV+E G S L L R
Sbjct 160 FLQEAAIMTRMRHEHVVRL---YGVVLDTKKIMLVSELATCG------SLLECLHKPALR 210
Query 66 RGVPPAAIRDFCERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFGA 125
P + D+ E++ + + L+R I L A R V P LVKI++FG
Sbjct 211 DSFPVHVLCDYAEQIAMG-----MSYLELQRLIHRDL---AARNVLVFSPKLVKISDFGL 262
Query 126 EEVLTIPSD 134
L I D
Sbjct 263 SRSLGIGED 271
> CE29073
Length=700
Score = 33.9 bits (76), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 3/43 (6%)
Query 46 EKVVAGWDDPRLSTLSALRRRGVPPAAIRDFCERVGVARRPST 88
E++ GW+D +T S+L +R A DFC+ +RRP T
Sbjct 88 EEIGVGWED---TTSSSLNKRMCKSATFHDFCDDSEYSRRPVT 127
> 7291302
Length=1426
Score = 31.6 bits (70), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/48 (37%), Positives = 27/48 (56%), Gaps = 7/48 (14%)
Query 101 YLDER-------APRRFAVKQPLLVKITNFGAEEVLTIPSDPKRPAAG 141
YL+E+ A R V+ P LVKIT+FG ++L+ S+ + A G
Sbjct 1053 YLEEKRLVHRDLAARNVLVQTPSLVKITDFGLAKLLSSDSNEYKAAGG 1100
> At5g14320
Length=197
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 29/84 (34%), Positives = 36/84 (42%), Gaps = 1/84 (1%)
Query 77 CERVGVARRPSTIQIELLERCIREYLDERAPRRFAVKQPLLVKITNFGAEEVLTIPSDPK 136
C RVG P+ +IE + I RA R+ V + KIT AEE L I D
Sbjct 60 CARVGGVEIPANKRIEYSLQYIHGIGRTRA-RQILVDLQMENKITKDMAEEELIILRDEV 118
Query 137 RPAAGSRQLAFTNELYIDAEDFME 160
LAF NEL I D ++
Sbjct 119 SKYMIEGDLAFVNELLICVVDVVQ 142
> SPAC343.11c
Length=1588
Score = 29.6 bits (65), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 19/62 (30%), Positives = 34/62 (54%), Gaps = 7/62 (11%)
Query 86 PSTIQI---ELLERCIREYLDERAPRRFAVKQPLLVKITNFGAEEVLTIPSDPKRPAAGS 142
PS+++I E+ CI +L ++K+ ++ +F A+ LT+PS+ K P A +
Sbjct 344 PSSLKILWKEVDYHCISSFLQSSNELASSLKK----QLPSFLAQTPLTLPSNTKTPPASA 399
Query 143 RQ 144
RQ
Sbjct 400 RQ 401
Lambda K H
0.321 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4562417408
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40