bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0278_orf2
Length=253
Score E
Sequences producing significant alignments: (Bits) Value
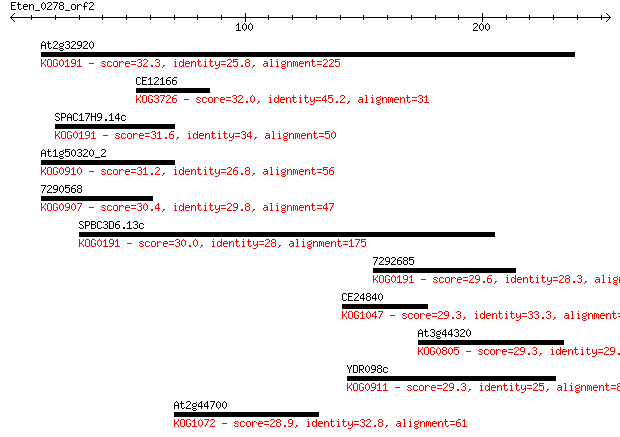
At2g32920 32.3 1.1
CE12166 32.0 1.3
SPAC17H9.14c 31.6 1.9
At1g50320_2 31.2 2.2
7290568 30.4 3.6
SPBC3D6.13c 30.0 4.5
7292685 29.6 6.7
CE24840 29.3 7.2
At3g44320 29.3 7.6
YDR098c 29.3 8.8
At2g44700 28.9 9.5
> At2g32920
Length=440
Score = 32.3 bits (72), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 58/231 (25%), Positives = 95/231 (41%), Gaps = 37/231 (16%)
Query 14 FQDKLAFVLLNKDSFPEAVKKYKVTSFPHLVVVRKDKKDEV-YKGAFEFQKLFDWLNVFS 72
Q K+ +N D + ++KV FP ++V DK Y+GA + + +
Sbjct 209 LQGKVKLGHVNCDVEQSIMSRFKVQGFPTILVFGPDKSSPYPYEGARSASAIESFASELV 268
Query 73 ETFVMGGGFSDTAPPAEIDPELQPWKVEPIPELTKQSSQDICFKKSNKFVCVMLAKR--G 130
E+ +A P E+ P +E K S ICF F+ +L + G
Sbjct 269 ES---------SAGPVEVTELTGPDVME-----KKCGSAAICFI---SFLPDILDSKAEG 311
Query 131 RQLTGEEQEMMESLKEDYEGHQDKGPDFRFMWIDLDTETEWAELFDIK--NTPTVVAVNP 188
R + EM+ S+ E ++ K P + FMW+ T+ + + ++ P +VA+N
Sbjct 312 RN---KYLEMLLSVAEKFK----KQP-YSFMWVAAVTQMDLEKRVNVGGYGYPAMVAMNV 363
Query 189 HKKVRF-LKLAADLPATKPHIRKMLGTISSGDARFKIVPQAKVPKFVDRKE 238
K V LK A +L ++ GT G+ VP P+ V KE
Sbjct 364 KKGVYAPLKSAFELQHLLEFVKDA-GTGGKGN-----VPMNGTPEIVKTKE 408
> CE12166
Length=725
Score = 32.0 bits (71), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 14/31 (45%), Positives = 21/31 (67%), Gaps = 1/31 (3%)
Query 54 VYKGAFEFQKLFDWLNVFSETFVMGGGFSDT 84
VY+ +FEFQ++F W +FSET + F D+
Sbjct 678 VYRMSFEFQRIFGW-RLFSETTRLRKNFQDS 707
> SPAC17H9.14c
Length=359
Score = 31.6 bits (70), Expect = 1.9, Method: Compositional matrix adjust.
Identities = 17/52 (32%), Positives = 29/52 (55%), Gaps = 2/52 (3%)
Query 20 FVLLNKDSFPEAVKKYKVTSFPHLVVVRKDKKD--EVYKGAFEFQKLFDWLN 69
V +N D F + + ++V SFP + KD KD E+Y+G + L +++N
Sbjct 195 IVKINADVFADIGRLHEVASFPTIKFFPKDDKDKPELYEGDRSLESLIEYIN 246
> At1g50320_2
Length=190
Score = 31.2 bits (69), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 15/58 (25%), Positives = 32/58 (55%), Gaps = 2/58 (3%)
Query 14 FQDKLAFVLLNKDSFPEAVKKYKVTSFPHLVVVRKDKK--DEVYKGAFEFQKLFDWLN 69
+ DKL V ++ D+ P+ + ++KV PH ++ + K+ +GA KL ++++
Sbjct 124 YGDKLTIVKIDHDANPKLIAEFKVYGLPHFILFKDGKEVPGSRREGAITKAKLKEYID 181
> 7290568
Length=107
Score = 30.4 bits (67), Expect = 3.6, Method: Compositional matrix adjust.
Identities = 14/47 (29%), Positives = 26/47 (55%), Gaps = 0/47 (0%)
Query 14 FQDKLAFVLLNKDSFPEAVKKYKVTSFPHLVVVRKDKKDEVYKGAFE 60
+ K + ++ D F E ++YKV S P V +R++++ + GA E
Sbjct 48 YSSKAVVLKIDVDKFEELTERYKVRSMPTFVFLRQNRRLASFAGADE 94
> SPBC3D6.13c
Length=726
Score = 30.0 bits (66), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 49/210 (23%), Positives = 82/210 (39%), Gaps = 52/210 (24%)
Query 30 EAVKKYKVTSFPHLVVVR------------KDKKDEVYKGAFEFQKLFDWLNVFSETFVM 77
+ KKY+V SFP L+VVR +D KD Y+ + K +WL V E
Sbjct 442 QIAKKYRVVSFPKLIVVRDGIASYYPAKMAQDNKD--YRRILGWMK-NNWLPVLPELRTS 498
Query 78 GGG--FSDTAPPA-EIDPELQPWKVEPIPELTKQSSQDICFKKSNKFVCVMLAKRGRQLT 134
F+D + ++PEL + + TK+++Q I + L + GR
Sbjct 499 NSKEIFNDESVVLFLLNPELDDF------DETKRTAQKIATE--------FLDEEGRTYQ 544
Query 135 ----------------GEEQEMMESLKEDYEGHQDKGPD-FRFMWIDLDTETEWAELFDI 177
EE+ +E+++ H + P RF W++ +W FDI
Sbjct 545 SNWQKETDKKNSLVNEAEEKNDLEAIEAAKNFHVNGKPSPTRFAWVNGKFWAQWLRKFDI 604
Query 178 ---KNTPTVVAVNPHKKVRFLKLAADLPAT 204
P V+ N + + + + A P +
Sbjct 605 DIEATGPRVIVYNAAQDIYWDETAKGTPIS 634
> 7292685
Length=335
Score = 29.6 bits (65), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 17/60 (28%), Positives = 27/60 (45%), Gaps = 0/60 (0%)
Query 154 KGPDFRFMWIDLDTETEWAELFDIKNTPTVVAVNPHKKVRFLKLAADLPATKPHIRKMLG 213
K P ID + F++K PT++ + KK+ A DL K ++ KM+G
Sbjct 132 KEPTVTISKIDCTQFRSICQDFEVKGYPTLLWIEDGKKIEKYSGARDLSTLKTYVEKMVG 191
> CE24840
Length=609
Score = 29.3 bits (64), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 12/36 (33%), Positives = 20/36 (55%), Gaps = 0/36 (0%)
Query 141 MESLKEDYEGHQDKGPDFRFMWIDLDTETEWAELFD 176
M +L Y+ Q K + +F W+ L ET+W+ + D
Sbjct 518 MPALTATYKLDQAKNAELKFSWLMLGLETKWSPIVD 553
> At3g44320
Length=346
Score = 29.3 bits (64), Expect = 7.6, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 31/66 (46%), Gaps = 5/66 (7%)
Query 173 ELFDIKNTPTVVAVNPHKKVRFLKLAA-----DLPATKPHIRKMLGTISSGDARFKIVPQ 227
E+ +KNT V+ V+P VR + + D PAT K + +S A+ + P+
Sbjct 6 EMSSVKNTTQVIGVDPSSTVRVTIVQSSTVYNDTPATLDKAEKFIVEAASKGAKLVLFPE 65
Query 228 AKVPKF 233
A + +
Sbjct 66 AFIGGY 71
> YDR098c
Length=285
Score = 29.3 bits (64), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 22/91 (24%), Positives = 43/91 (47%), Gaps = 5/91 (5%)
Query 143 SLKEDYEG--HQDKGPDFRFMWIDLDTETEWAELFDIKNTPTVVAVNPHKKVRFLKLA-A 199
+LK+ +E ++ + F+ ID D +E +ELF+I P + + HK +L+ A
Sbjct 74 ALKQVFEAISNEPSNSNVSFLSIDADENSEISELFEISAVPYFIII--HKGTILKELSGA 131
Query 200 DLPATKPHIRKMLGTISSGDARFKIVPQAKV 230
D + +++SG ++ + A V
Sbjct 132 DPKEYVSLLEDCKNSVNSGSSQTHTMENANV 162
> At2g44700
Length=368
Score = 28.9 bits (63), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 5/64 (7%)
Query 70 VFSETFVMGGGFSDTAPPAEIDPELQPWKVEPIPELTKQSSQDICFKKSNKF---VCVML 126
V + +V+GG D D Q W+ PIP+ K+S + IC K + F VC +
Sbjct 174 VGGKVYVIGGYQDDEIAAESFDLNTQTWEAAPIPD-EKESHRWIC-KANVSFDRKVCALR 231
Query 127 AKRG 130
++ G
Sbjct 232 SREG 235
Lambda K H
0.318 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5223052002
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40