bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0277_orf2
Length=115
Score E
Sequences producing significant alignments: (Bits) Value
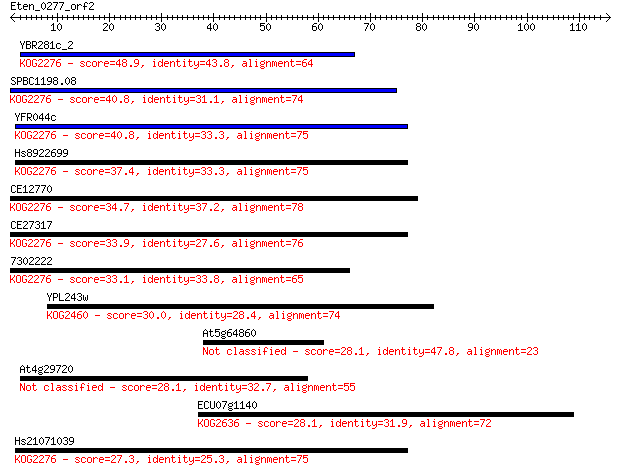
YBR281c_2 48.9 3e-06
SPBC1198.08 40.8 6e-04
YFR044c 40.8 6e-04
Hs8922699 37.4 0.007
CE12770 34.7 0.050
CE27317 33.9 0.076
7302222 33.1 0.14
YPL243w 30.0 1.0
At5g64860 28.1 4.5
At4g29720 28.1 4.5
ECU07g1140 28.1 4.7
Hs21071039 27.3 6.1
> YBR281c_2
Length=452
Score = 48.9 bits (115), Expect = 3e-06, Method: Composition-based stats.
Identities = 28/64 (43%), Positives = 34/64 (53%), Gaps = 14/64 (21%)
Query 3 WGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPNER 62
W VEPL VREGGS+ + L F D AV Q+P GQ++DN HL NE
Sbjct 387 WDVEPLLVREGGSISCLRMLERIF-------------DAPAV-QIPCGQSTDNGHLANEN 432
Query 63 LGIR 66
L I+
Sbjct 433 LRIK 436
> SPBC1198.08
Length=474
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 14/74 (18%)
Query 1 QVWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPN 60
+V+G+ P +VREGGS+P + ++ V LP G+ D AH N
Sbjct 402 RVYGITPDFVREGGSIPVTVTFEQSLKK--------------NVLLLPMGRGDDGAHSIN 447
Query 61 ERLGIRQVEKAAKL 74
E+L + K KL
Sbjct 448 EKLDLDNFLKGIKL 461
> YFR044c
Length=481
Score = 40.8 bits (94), Expect = 6e-04, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 32/75 (42%), Gaps = 14/75 (18%)
Query 2 VWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPNE 61
V+GV+P + REGGS+P DA +V LP G+ D AH NE
Sbjct 409 VYGVDPDFTREGGSIPITLTFQDAL--------------NTSVLLLPMGRGDDGAHSINE 454
Query 62 RLGIRQVEKAAKLTA 76
+L I K A
Sbjct 455 KLDISNFVGGMKTMA 469
> Hs8922699
Length=475
Score = 37.4 bits (85), Expect = 0.007, Method: Composition-based stats.
Identities = 25/75 (33%), Positives = 32/75 (42%), Gaps = 14/75 (18%)
Query 2 VWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPNE 61
V+GVEP REGGS+P +A + V LP G A D AH NE
Sbjct 404 VFGVEPDLTREGGSIPVTLTFQEATGK--------------NVMLLPVGSADDGAHSQNE 449
Query 62 RLGIRQVEKAAKLTA 76
+L + K+ A
Sbjct 450 KLNRYNYIEGTKMLA 464
> CE12770
Length=473
Score = 34.7 bits (78), Expect = 0.050, Method: Composition-based stats.
Identities = 29/79 (36%), Positives = 36/79 (45%), Gaps = 15/79 (18%)
Query 1 QVWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPN 60
+V GVEP +REG S+P + F+ L G +V LP G A D AH N
Sbjct 403 RVHGVEPDRIREGCSIP----ITLTFQELTGK----------SVLLLPIGAADDMAHSQN 448
Query 61 ERLGI-RQVEKAAKLTANI 78
E+ I VE L A I
Sbjct 449 EKNNIWNYVEGVKTLLAYI 467
> CE27317
Length=461
Score = 33.9 bits (76), Expect = 0.076, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 33/76 (43%), Gaps = 14/76 (18%)
Query 1 QVWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPN 60
+++G+ P + REGGS+P + D + V LP G + D AH N
Sbjct 391 KLYGMTPDFTREGGSIPVTLTIQDLTKS--------------PVMLLPIGASDDMAHSQN 436
Query 61 ERLGIRQVEKAAKLTA 76
E++ K K+ A
Sbjct 437 EKINRDNFVKGMKVLA 452
> 7302222
Length=462
Score = 33.1 bits (74), Expect = 0.14, Method: Composition-based stats.
Identities = 22/65 (33%), Positives = 29/65 (44%), Gaps = 14/65 (21%)
Query 1 QVWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPN 60
V+ VEP REGGS+P L +A G + + V P G D AH N
Sbjct 404 HVFNVEPDMTREGGSIPVTLTLQEA-----------TGKNVILV---PVGACDDGAHSQN 449
Query 61 ERLGI 65
E++ I
Sbjct 450 EKIDI 454
> YPL243w
Length=599
Score = 30.0 bits (66), Expect = 1.0, Method: Composition-based stats.
Identities = 21/74 (28%), Positives = 29/74 (39%), Gaps = 14/74 (18%)
Query 8 LYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPNERLGIRQ 67
LY +G M A+ DA+RRL L + + DE+ LP L +
Sbjct 426 LYQSKGRYMEALALYVDAYRRLENKLSEIESLDEIL--------------LPANLLSLNS 471
Query 68 VEKAAKLTANIGSS 81
V K N G+S
Sbjct 472 VRSLQKRIENGGNS 485
> At5g64860
Length=576
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 11/23 (47%), Positives = 15/23 (65%), Gaps = 0/23 (0%)
Query 38 GSDEVAVCQLPFGQASDNAHLPN 60
G+ +AV Q FG +DN HLP+
Sbjct 437 GAPGMAVLQFAFGGGADNPHLPH 459
> At4g29720
Length=533
Score = 28.1 bits (61), Expect = 4.5, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 29/64 (45%), Gaps = 9/64 (14%)
Query 3 WGVEPL------YVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPF---GQAS 53
WG +PL YV G S + +A+ ++ +GQ G D+ V +L G+A+
Sbjct 444 WGSDPLFRGSYSYVAVGSSGDDLDAMAEPLPKINKKVGQVNGHDQAKVHELQVMFAGEAT 503
Query 54 DNAH 57
H
Sbjct 504 HRTH 507
> ECU07g1140
Length=370
Score = 28.1 bits (61), Expect = 4.7, Method: Composition-based stats.
Identities = 23/73 (31%), Positives = 36/73 (49%), Gaps = 6/73 (8%)
Query 37 QGSDEVAVCQLPFGQASDNAHLPNERLGIRQVEKAAKLTANIGSSRGP-SMSAFLRALCG 95
QG +EV C +P +A L + L I +EK K + ++ S R + S+ R LC
Sbjct 221 QGKEEVLYCGMPVSEAES---LVLKSLAI--LEKERKHSISLLSRRKKQTKSSVPRWLCR 275
Query 96 TLELEICLTPDIC 108
+L+I +IC
Sbjct 276 RKDLDIAFECEIC 288
> Hs21071039
Length=507
Score = 27.3 bits (59), Expect = 6.1, Method: Composition-based stats.
Identities = 19/75 (25%), Positives = 32/75 (42%), Gaps = 14/75 (18%)
Query 2 VWGVEPLYVREGGSMPAIPFLADAFRRLPGPLGQCQGSDEVAVCQLPFGQASDNAHLPNE 61
V+G EP +R+G ++P +A F+ + +V +P G D H NE
Sbjct 437 VFGTEPDMIRDGSTIP----IAKMFQEIV----------HKSVVLIPLGAVDDGEHSQNE 482
Query 62 RLGIRQVEKAAKLTA 76
++ + KL A
Sbjct 483 KINRWNYIEGTKLFA 497
Lambda K H
0.322 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1178471386
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40