bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0265_orf1
Length=188
Score E
Sequences producing significant alignments: (Bits) Value
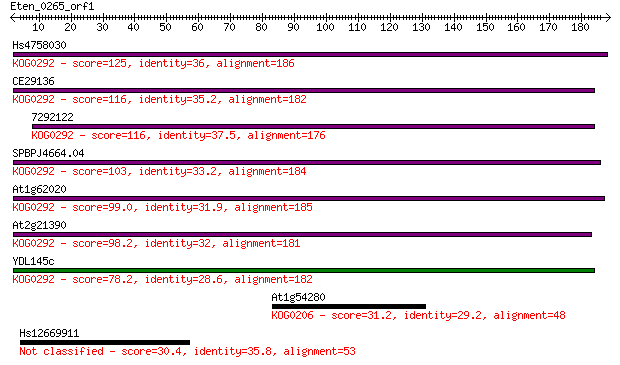
Hs4758030 125 4e-29
CE29136 116 2e-26
7292122 116 3e-26
SPBPJ4664.04 103 2e-22
At1g62020 99.0 5e-21
At2g21390 98.2 8e-21
YDL145c 78.2 9e-15
At1g54280 31.2 1.4
Hs12669911 30.4 2.0
> Hs4758030
Length=1224
Score = 125 bits (315), Expect = 4e-29, Method: Composition-based stats.
Identities = 67/189 (35%), Positives = 107/189 (56%), Gaps = 10/189 (5%)
Query 2 VAQNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGES---DAQRNLELVAYFSCCRLQPS 58
V N++E + +++ ICR YI+G+ +ET R L + +R E+ AYF+ LQP
Sbjct 1040 VVDNKQEIAEAQQLITICREYIVGLSVETERKKLPKETLEQQKRICEMAAYFTHSNLQPV 1099
Query 59 HSFLVLRRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTD 118
H LVLR A+++ +K +NF TAA+FARRLL L PE + +++L CE+ TD
Sbjct 1100 HMILVLRTALNLFFKLKNFKTAATFARRLLE-----LGPKPEVAQQTRKILSACEKNPTD 1154
Query 119 AININYEPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGAR 178
A +NY+ +I C ++ + G PV +C GA + +G +CRV + E+G
Sbjct 1155 AYQLNYDMHNPFDI--CAASYRPIYRGKPVEKCPLSGACYSPEFKGQICRVTTVTEIGKD 1212
Query 179 VVGIQFLPV 187
V+G++ P+
Sbjct 1213 VIGLRISPL 1221
> CE29136
Length=1232
Score = 116 bits (291), Expect = 2e-26, Method: Composition-based stats.
Identities = 64/185 (34%), Positives = 103/185 (55%), Gaps = 10/185 (5%)
Query 2 VAQNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGES---DAQRNLELVAYFSCCRLQPS 58
V +++E + +++ I R Y+ + LET R L ++ DA+RN EL AYF+ LQP
Sbjct 1049 VVSSKQEVAEAEQLITITREYLAALLLETYRKDLPKTNLEDAKRNAELAAYFTHFELQPM 1108
Query 59 HSFLVLRRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTD 118
H L LR A++ +K + T AS +RLL L PE A+ ++VL E+ TD
Sbjct 1109 HRILTLRSAINTFFKMKQMKTCASLCKRLLE-----LAPKPEVAAQIRKVLTAAEKDNTD 1163
Query 119 AININYEPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGAR 178
A + Y+ E ++C+ L G P+ +C +CGA + L+G +C VC++AE+G
Sbjct 1164 AHQLTYD--EHNPFVVCSRQFVPLYRGRPLCKCPYCGASYSEGLEGEVCNVCQVAEVGKN 1221
Query 179 VVGIQ 183
V+G++
Sbjct 1222 VLGLR 1226
> 7292122
Length=1234
Score = 116 bits (290), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 66/179 (36%), Positives = 101/179 (56%), Gaps = 13/179 (7%)
Query 8 EEQQLLEMLDICRTYIIGMRLETSRLMLGES---DAQRNLELVAYFSCCRLQPSHSFLVL 64
E QQLL IC YI+G+++ET R + +S + +R E+ AYF+ C+LQP H L L
Sbjct 1059 EAQQLLR---ICAEYIVGLKMETVRKGMPKSTLEEQKRLCEMAAYFTHCKLQPVHQILTL 1115
Query 65 RRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTDAININY 124
R A+++ +K +N+ TAASFARRLL L P+ + +++L CE D + Y
Sbjct 1116 RTALNMFFKLKNYKTAASFARRLLE-----LAPRPDVAQQVRKILQACEVNPVDEHQLQY 1170
Query 125 EPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGARVVGIQ 183
E E +C + L G P V C FC + Q +G+LC VCE++++G +G++
Sbjct 1171 E--EFNPFTICGISWKPLYRGKPEVTCPFCSSSFDPQFKGNLCTVCEVSQIGKDSIGLR 1227
> SPBPJ4664.04
Length=1207
Score = 103 bits (257), Expect = 2e-22, Method: Composition-based stats.
Identities = 61/184 (33%), Positives = 93/184 (50%), Gaps = 7/184 (3%)
Query 2 VAQNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGESDAQRNLELVAYFSCCRLQPSHSF 61
VA ++EE ++ ++D C YI+ + E R LGE D +R LEL YF+ LQP HS
Sbjct 1030 VANSEEEADEISALIDECCRYIVALSCELERRRLGEEDTKRALELSYYFASADLQPMHSI 1089
Query 62 LVLRRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTDAIN 121
+ LR A++ + K +N+ +A+ +LL SG P A A R + L ++ DA
Sbjct 1090 IALRLAINASHKLKNYKSASFLGNKLLQLAESG----PAAEA-ANRAITLGDRNPHDAFE 1144
Query 122 INYEPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGARVVG 181
I Y+P I C T+T + G C CGAV + +C VC++ +G + G
Sbjct 1145 IEYDPHVEMRI--CPKTLTPVYSGDDFDVCSVCGAVYHKGYVNEVCTVCDVGGIGQKGTG 1202
Query 182 IQFL 185
+F
Sbjct 1203 RRFF 1206
> At1g62020
Length=1216
Score = 99.0 bits (245), Expect = 5e-21, Method: Composition-based stats.
Identities = 59/185 (31%), Positives = 97/185 (52%), Gaps = 8/185 (4%)
Query 2 VAQNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGESDAQRNLELVAYFSCCRLQPSHSF 61
V + + E ++ E++ I + Y++G+++E R + + D R EL AYF+ C LQ H
Sbjct 1036 VVETRREVDEVKELIVIVKEYVLGLQMELKRREM-KDDPVRQQELAAYFTHCNLQTPHLR 1094
Query 62 LVLRRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTDAIN 121
L L AM V +KA+N TA++FARRLL + + A++V+ E+ TD
Sbjct 1095 LALLSAMGVCYKAKNLATASNFARRLLETS-----PVDSQAKMARQVVQAAERNMTDETK 1149
Query 122 INYEPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGARVVG 181
+NY+ ++C ST + G V C +C A +G++C VC++A +GA G
Sbjct 1150 LNYD--FRNPFVVCGSTYVPIYRGQKDVSCPYCTARFVPNQEGNICTVCDLAVIGADASG 1207
Query 182 IQFLP 186
+ P
Sbjct 1208 LLCSP 1212
> At2g21390
Length=1218
Score = 98.2 bits (243), Expect = 8e-21, Method: Composition-based stats.
Identities = 58/181 (32%), Positives = 97/181 (53%), Gaps = 7/181 (3%)
Query 2 VAQNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGESDAQRNLELVAYFSCCRLQPSHSF 61
V +++ E ++ E++ I + Y++G++LE R + + D R EL AYF+ C+LQ H
Sbjct 1037 VVESRREVDEVKELVIIVKEYVLGLQLELKRREM-KDDPVRQQELAAYFTHCKLQTPHLR 1095
Query 62 LVLRRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTDAIN 121
L AM+V +K++N TAA FAR LL + + A++V+ E+ TDA
Sbjct 1096 LAYFSAMTVCYKSKNMATAAHFARSLLDTNPT----IESQARTARQVMQAAERNMTDATT 1151
Query 122 INYEPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGARVVG 181
+NY+ ++C ST + G V C +C A +G++C VC++A +GA G
Sbjct 1152 LNYD--FRNPFVICGSTYVPIYKGQKDVACPYCTARFVPSQEGNICSVCDLAVIGADASG 1209
Query 182 I 182
+
Sbjct 1210 L 1210
> YDL145c
Length=1201
Score = 78.2 bits (191), Expect = 9e-15, Method: Composition-based stats.
Identities = 52/182 (28%), Positives = 92/182 (50%), Gaps = 7/182 (3%)
Query 2 VAQNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGESDAQRNLELVAYFSCCRLQPSHSF 61
+ + E+E+ ++L+ R YI+G+ +E R L E + R LEL AYF+ +L P H
Sbjct 1024 MVDDAEDEKLAHKILETAREYILGLSIELERRSLKEGNTVRMLELAAYFTKAKLSPIHRT 1083
Query 62 LVLRRAMSVAWKAQNFITAASFARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTDAIN 121
L+ AMS +K +NF+ A+ FA L SG + +A+++ + +DAI
Sbjct 1084 NALQVAMSQHFKHKNFLQASYFAGEFLKIISSGPRA-----EQARKIKNKADSMASDAIP 1138
Query 122 INYEPGEAENILLCTSTMTRLNPGTPVVRCGFCGAVAQQQLQGSLCRVCEIAELGARVVG 181
I+++P +I C +T + TP V G+ + + R+ I+++GA G
Sbjct 1139 IDFDPYAKFDI--CAATYKPIYEDTPSVSDPLTGSKYVITEKDKIDRIAMISKIGAPASG 1196
Query 182 IQ 183
++
Sbjct 1197 LR 1198
> At1g54280
Length=1240
Score = 31.2 bits (69), Expect = 1.4, Method: Composition-based stats.
Identities = 14/48 (29%), Positives = 28/48 (58%), Gaps = 0/48 (0%)
Query 83 FARRLLSGTYSGLKGAPEELAKAKRVLLLCEQKGTDAININYEPGEAE 130
F + ++GT G++ + ELA AK++ + E+KG + N++ G +
Sbjct 438 FLKCSIAGTSYGVRASEVELAAAKQMAMDLEEKGEEVANLSMNKGRTQ 485
> Hs12669911
Length=437
Score = 30.4 bits (67), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 19/53 (35%), Positives = 30/53 (56%), Gaps = 12/53 (22%)
Query 4 QNQEEEQQLLEMLDICRTYIIGMRLETSRLMLGESDAQRNLELVAYFSCCRLQ 56
Q QE EQQL +++IC T + RL+ ++D+QR +AY +C L+
Sbjct 212 QLQESEQQLDHLMNICTTQL--------RLLSEDTDSQR----LAYVTCQDLR 252
Lambda K H
0.323 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3094507556
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40