bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
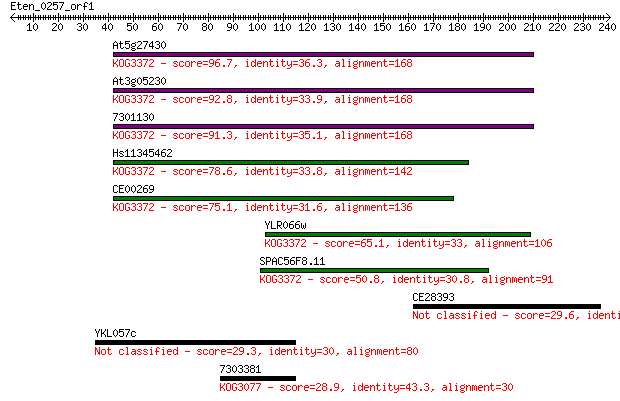
Query= Eten_0257_orf1
Length=240
Score E
Sequences producing significant alignments: (Bits) Value
At5g27430 96.7 4e-20
At3g05230 92.8 6e-19
7301130 91.3 1e-18
Hs11345462 78.6 1e-14
CE00269 75.1 1e-13
YLR066w 65.1 1e-10
SPAC56F8.11 50.8 2e-06
CE28393 29.6 5.3
YKL057c 29.3 8.3
7303381 28.9 9.9
> At5g27430
Length=167
Score = 96.7 bits (239), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 61/171 (35%), Positives = 91/171 (53%), Gaps = 8/171 (4%)
Query 42 MESYLNRANAVFCALMVSLSVLAIGNVISSY---FLMGPISGSIAVKDVYGFSYNYALNG 98
M S+ RANA+ L ++++LA I+S+ F S I + ++ F N
Sbjct 1 MHSFGYRANAL---LTFAVTILAFICAIASFSDNFSNQNPSAQIQILNINWFQKQPHGN- 56
Query 99 DQAVLSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIMDDGVAVIDWP 158
D+ L+L+I ADL+ LF WN KQV+ FV AEYET ++ NQV ++D II + A W
Sbjct 57 DEVSLTLNITADLQSLFTWNTKQVFAFVAAEYETSKNALNQVSLWDAIIPEKEHAKF-WI 115
Query 159 NTPAKYHFRDKGRGLRGREVTVKLQVVYHPIVGRMYTQTIATNTFRMPGQY 209
KY F D+G LRG++ + L P G+M+ I + +R+P Y
Sbjct 116 QISNKYRFIDQGHNLRGKDFNLTLHWHVMPKTGKMFADKIVMSGYRLPNAY 166
> At3g05230
Length=167
Score = 92.8 bits (229), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 57/168 (33%), Positives = 85/168 (50%), Gaps = 2/168 (1%)
Query 42 MESYLNRANAVFCALMVSLSVLAIGNVISSYFLMGPISGSIAVKDVYGFSYNYALNGDQA 101
M ++ RANA+ + +L+ + S F S I + ++ F N D+
Sbjct 1 MHTFGYRANALLTFAVTALAFICAIASFSDKFSNQNPSAEIQILNINRFKKQSHGN-DEV 59
Query 102 VLSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIMDDGVAVIDWPNTP 161
L+LDI ADL+ LF WN KQV++FV AEYETP++ NQV ++D II A
Sbjct 60 SLTLDISADLQSLFTWNTKQVFVFVAAEYETPKNSLNQVSLWDAIIPAKEHAKFR-IQVS 118
Query 162 AKYHFRDKGRGLRGREVTVKLQVVYHPIVGRMYTQTIATNTFRMPGQY 209
KY F D+G+ LRG++ + L P G+M+ I + +P Y
Sbjct 119 NKYRFIDQGQNLRGKDFNLTLHWHVMPKTGKMFADKIVLPGYSLPDAY 166
> 7301130
Length=179
Score = 91.3 bits (225), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 59/177 (33%), Positives = 92/177 (51%), Gaps = 12/177 (6%)
Query 42 MESYLNRANAVFCALMVSLSVLAIGNVISSYFLMGPISGSI-----AVKDV--YGFSYNY 94
M + L R NA + L+ L +S+ FL +I VK+V YG S
Sbjct 1 MHTVLTRGNATVAYTLSVLACLTFSCFLSTVFLDYRTDANINTVRVLVKNVPDYGASRE- 59
Query 95 ALNGDQAVLSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIMDDGVAV 154
D ++ D++ +L G+F WN KQ++L++ AEY+TP + NQVV++D+II+ AV
Sbjct 60 --KHDLGFVTFDLQTNLTGIFNWNVKQLFLYLTAEYQTPANQLNQVVLWDKIILRGDNAV 117
Query 155 IDWPNTPAKYHFRDKGRGLR-GREVTVKLQVVYHPIVGRMYT-QTIATNTFRMPGQY 209
+D+ N KY+F D G GL+ R V++ L P G + + Q + F+ P Y
Sbjct 118 LDFKNMNTKYYFWDDGNGLKDNRNVSLYLSWNIIPNAGLLPSVQATGKHLFKFPADY 174
> Hs11345462
Length=180
Score = 78.6 bits (192), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 48/148 (32%), Positives = 78/148 (52%), Gaps = 7/148 (4%)
Query 42 MESYLNRANAVFCALMVSLSVLAIGNVISSYFLMGPIS-----GSIAVKDVYGFSYNYAL 96
M + L+RAN++F + ++ L G I++ F + I +K+V F+
Sbjct 1 MNTVLSRANSLFAFSLSVMAALTFGCFITTAFKDRSVPVRLHVSRIMLKNVEDFTGPRE- 59
Query 97 NGDQAVLSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIMDDGVAVID 156
D ++ DI ADL +F WN KQ++L++ AEY T + NQVV++D+I++ +
Sbjct 60 RSDLGFITFDITADLENIFDWNVKQLFLYLSAEYSTKNNALNQVVLWDKIVLRGDNPKLL 119
Query 157 WPNTPAKYHFRDKGRGLRG-REVTVKLQ 183
+ KY F D G GL+G R VT+ L
Sbjct 120 LKDMKTKYFFFDDGNGLKGNRNVTLTLS 147
> CE00269
Length=180
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/140 (30%), Positives = 76/140 (54%), Gaps = 4/140 (2%)
Query 42 MESYLNRANAVFCALMVSLSVLAIGNVISSYFLMGPISGSIAVKDVYGFSY-NYALN--- 97
M + L+RANA+ + ++ + +S+ FL + + V DV + +YA +
Sbjct 1 MHNLLSRANALLAFTLWVMAAVTAACFLSTVFLDYTVPTKLTVNDVKVRNVVDYATDEQQ 60
Query 98 GDQAVLSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIMDDGVAVIDW 157
D A L+ ++K D +F WN KQ+++++VAEY++ + NQVV++DRI+ V+D
Sbjct 61 ADLATLNFNLKVDFSKIFNWNVKQLFVYLVAEYKSKVNEVNQVVLWDRIVERADRVVMDE 120
Query 158 PNTPAKYHFRDKGRGLRGRE 177
+KY+F D G L +
Sbjct 121 IGVKSKYYFLDDGTNLLNHK 140
> YLR066w
Length=184
Score = 65.1 bits (157), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 57/108 (52%), Gaps = 2/108 (1%)
Query 103 LSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIMDDGVAVIDWPNTPA 162
+ D+ DL LF WN KQV++++ AEY + + T++V +D+II AVID + +
Sbjct 74 IKFDLNTDLTPLFNWNTKQVFVYLTAEYNSTEKITSEVTFWDKIIKSKDDAVIDVNDLRS 133
Query 163 KYHFRDKGRG-LRGREVTVKLQVVYHPIVGRM-YTQTIATNTFRMPGQ 208
KY D G G+++ KL P VG + Y +T+ T + +
Sbjct 134 KYSIWDIEDGKFEGKDLVFKLHWNVQPWVGLLTYGETVGNYTLTVENK 181
> SPAC56F8.11
Length=185
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 28/94 (29%), Positives = 47/94 (50%), Gaps = 3/94 (3%)
Query 101 AVLSLDIKADLRGLFQWNAKQVYLFVVAEYETPQHPTNQVVVFDRIIM---DDGVAVIDW 157
A + ++ ADL L+ WN K V +++VA Y T +H NQVVV+D+I+ + + + D
Sbjct 69 AQVKFNMDADLSELWDWNTKHVVVYLVASYSTEKHEKNQVVVWDKILSSPEESKMFMKDT 128
Query 158 PNTPAKYHFRDKGRGLRGREVTVKLQVVYHPIVG 191
+ + F + G+ T L P +G
Sbjct 129 LSNIQAHPFNEYSNQFEGKNATYTLHWTVSPKMG 162
> CE28393
Length=1292
Score = 29.6 bits (65), Expect = 5.3, Method: Composition-based stats.
Identities = 18/75 (24%), Positives = 31/75 (41%), Gaps = 1/75 (1%)
Query 162 AKYHFRDKGRGLRGREVTVKLQVVYHPIVGRMYTQTIATNTFRMPGQYDRVTHKQQQEQQ 221
AK+ + +R RE+++ + VYH G YT + + M G H+ Q
Sbjct 187 AKFFAEKEENHIRTRELSLSVHPVYHQSNGDAYTSFGMSRSKSM-GSVAHEAHQTSNNQH 245
Query 222 QQQAREAAMAAAARP 236
+ A + +RP
Sbjct 246 SSETHGTASSTQSRP 260
> YKL057c
Length=1037
Score = 29.3 bits (64), Expect = 8.3, Method: Composition-based stats.
Identities = 24/91 (26%), Positives = 38/91 (41%), Gaps = 15/91 (16%)
Query 35 KNLAAGKMESYLNRANAVFCALMVSLSVLAIGNVISSYFLMGPISGSIAVKDVYGFSYN- 93
+NL +G E + ++C M++ S A V+ Y LM I + Y N
Sbjct 951 QNLRSGDWECF----KKLYCFRMLNKSERAAAEVLYQYILMQADLDVIRKRKCYLMVINV 1006
Query 94 ----------YALNGDQAVLSLDIKADLRGL 114
+ LNG + V D++ +LRGL
Sbjct 1007 LSSFDSAYDQWILNGSKVVTLTDLRDELRGL 1037
> 7303381
Length=288
Score = 28.9 bits (63), Expect = 9.9, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 85 KDVYGFSYNYALNGDQAVLSLDIKADLRGL 114
K +Y F++ + L DQ VLSL++ DL L
Sbjct 162 KQLYRFTFRFGLEPDQRVLSLEMAIDLWKL 191
Lambda K H
0.324 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4811924610
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40