bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
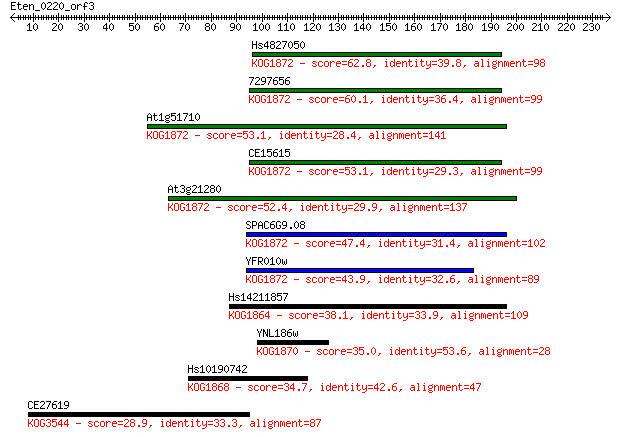
Query= Eten_0220_orf3
Length=236
Score E
Sequences producing significant alignments: (Bits) Value
Hs4827050 62.8 6e-10
7297656 60.1 4e-09
At1g51710 53.1 5e-07
CE15615 53.1 5e-07
At3g21280 52.4 9e-07
SPAC6G9.08 47.4 2e-05
YFR010w 43.9 3e-04
Hs14211857 38.1 0.015
YNL186w 35.0 0.12
Hs10190742 34.7 0.18
CE27619 28.9 9.0
> Hs4827050
Length=494
Score = 62.8 bits (151), Expect = 6e-10, Method: Compositional matrix adjust.
Identities = 39/99 (39%), Positives = 49/99 (49%), Gaps = 36/99 (36%)
Query 96 GTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRR 155
G Y L +V+THQGR + SGHYV W KR+
Sbjct 416 GYYDLQAVLTHQGRSSSSGHYVSWV--------------------------------KRK 443
Query 156 KNVDMWRKFDDDKVSEV-PWDSIDLAGGRSDYHVAYLLL 193
+ D W KFDDDKVS V P D + L+GG D+H+AY+LL
Sbjct 444 Q--DEWIKFDDDKVSIVTPEDILRLSGG-GDWHIAYVLL 479
> 7297656
Length=475
Score = 60.1 bits (144), Expect = 4e-09, Method: Compositional matrix adjust.
Identities = 36/99 (36%), Positives = 46/99 (46%), Gaps = 34/99 (34%)
Query 95 TGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKR 154
+G Y L +V+TH+GR + SGHYV W R+ SG
Sbjct 402 SGYYTLQAVLTHKGRSSSSGHYVAWVRS-------------------------SG----- 431
Query 155 RKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLL 193
D+W KFDDD+VS V D I G D+H AY+LL
Sbjct 432 ----DVWFKFDDDEVSAVATDEILRLSGGGDWHCAYVLL 466
> At1g51710
Length=490
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 59/141 (41%), Gaps = 48/141 (34%)
Query 55 AATNGSGAAHSASAPAAEAAEAEAKKKETELLRLAGKPCPTGTYQLLSVVTHQGRYADSG 114
A+ NGSG + + + ++E KET + TG Y L++V+TH+GR ADSG
Sbjct 392 ASANGSGESSTVNPQEGTSSE-----KETHM---------TGIYDLVAVLTHKGRSADSG 437
Query 115 HYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRRKNVDMWRKFDDDKVSEVPW 174
HYV W + SG W ++DDD S
Sbjct 438 HYVAWVK------------------------QESGK----------WIQYDDDNPSMQRE 463
Query 175 DSIDLAGGRSDYHVAYLLLMK 195
+ I G D+H+AY+ + K
Sbjct 464 EDITKLSGGGDWHMAYITMYK 484
> CE15615
Length=489
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 29/99 (29%), Positives = 42/99 (42%), Gaps = 33/99 (33%)
Query 95 TGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKR 154
+G Y L ++TH+GR + GHYV W R+ + G
Sbjct 389 SGFYDLKGIITHKGRSSQDGHYVAWMRSSEDGK--------------------------- 421
Query 155 RKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLL 193
WR FDD+ V+ V ++I G D+H AY+LL
Sbjct 422 ------WRLFDDEHVTVVDEEAILKTSGGGDWHSAYVLL 454
> At3g21280
Length=488
Score = 52.4 bits (124), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 41/141 (29%), Positives = 63/141 (44%), Gaps = 39/141 (27%)
Query 63 AHSASAPAAEAAEAEAKKKETELLRL---AGKPCPTGTYQLLSVVTHQGRYADSGHYVGW 119
A +S + E++ + ++ LL+ P TG Y L+SV+TH+GR ADSGHYV W
Sbjct 381 AEGSSNQSGESSTGDQQEVSISLLQTFATGASPHMTGIYDLVSVLTHKGRSADSGHYVAW 440
Query 120 ARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAKRRKNVDMWRKFDDDKVS-EVPWDSID 178
+ SG W ++DD S + D I
Sbjct 441 VK------------------------QESGK----------WVQYDDANTSLQRGEDIIK 466
Query 179 LAGGRSDYHVAYLLLMKHILV 199
L+GG D+H+AY+++ K L+
Sbjct 467 LSGG-GDWHMAYIVMYKARLI 486
> SPAC6G9.08
Length=467
Score = 47.4 bits (111), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 32/102 (31%), Positives = 43/102 (42%), Gaps = 32/102 (31%)
Query 94 PTGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAK 153
PTG Y L+ V++H G A SGHY W R S ++ E
Sbjct 393 PTGLYDLVGVLSHAGASASSGHYQAWIR-------------NSNNRAE------------ 427
Query 154 RRKNVDMWRKFDDDKVSEVPWDSIDLAGGRSDYHVAYLLLMK 195
W +F+D KVS VP + I+ G + AY+LL K
Sbjct 428 -------WFRFNDAKVSIVPAEKIETLDGGGEADSAYILLYK 462
> YFR010w
Length=499
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 33/90 (36%)
Query 94 PTGTYQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQSEDKEEEEPAAPSGPAAK 153
P+ Y L+ V+THQG ++SGHY + R D+ +E
Sbjct 426 PSCVYNLIGVITHQGANSESGHYQAFIR----------------DELDE----------- 458
Query 154 RRKNVDMWRKFDDDKVSEVPWDSID-LAGG 182
+ W KF+DDKVS V + I+ LAGG
Sbjct 459 -----NKWYKFNDDKVSVVEKEKIESLAGG 483
> Hs14211857
Length=397
Score = 38.1 bits (87), Expect = 0.015, Method: Compositional matrix adjust.
Identities = 37/129 (28%), Positives = 52/129 (40%), Gaps = 20/129 (15%)
Query 87 RLAGKPCPTGT----------YQLLSVVTHQGRYADSGHYVGWARAGDAGTLQGPPTAQS 136
LA K P+GT Y L SVV H G ++SGHY +AR + QS
Sbjct 174 NLAKKLKPSGTDEASCTKLVPYLLSSVVVHSGISSESGHYYSYARNITSTDSSYQMYHQS 233
Query 137 E-------DKEEEEPAAPSGPAAKRRKNVDM---WRKFDDDKVSEVPWDSIDLAGGRSDY 186
E +PS + +N +M W F+D +V+ + S+ R
Sbjct 234 EALALASSQSHLLGRDSPSAVFEQDLENKEMSKEWFLFNDSRVTFTSFQSVQKITSRFPK 293
Query 187 HVAYLLLMK 195
AY+LL K
Sbjct 294 DTAYVLLYK 302
> YNL186w
Length=792
Score = 35.0 bits (79), Expect = 0.12, Method: Composition-based stats.
Identities = 15/28 (53%), Positives = 18/28 (64%), Gaps = 0/28 (0%)
Query 98 YQLLSVVTHQGRYADSGHYVGWARAGDA 125
YQLLSVV H+GR SGHY+ + D
Sbjct 674 YQLLSVVVHEGRSLSSGHYIAHCKQPDG 701
> Hs10190742
Length=922
Score = 34.7 bits (78), Expect = 0.18, Method: Composition-based stats.
Identities = 20/47 (42%), Positives = 25/47 (53%), Gaps = 4/47 (8%)
Query 71 AEAAEAEAKKKETELLRLAGKPCPTGTYQLLSVVTHQGRYADSGHYV 117
AE EAK EL R P Y+L+SVV+H G +SGHY+
Sbjct 800 AENTRGEAK----ELTRNVKMGDPLQAYRLISVVSHIGSSPNSGHYI 842
> CE27619
Length=431
Score = 28.9 bits (63), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 29/89 (32%), Positives = 34/89 (38%), Gaps = 6/89 (6%)
Query 8 VALRREREASSGGAEKTTNGTASTESGAPAADGAAAATAAPEAAAVPAATNGS-GAAHSA 66
+ R R A G + NG S SGAP G A P AA PA T G G A
Sbjct 70 IPASRNRTARQAGYDAGVNGGGSEASGAP---GNCEACCLPGAAG-PAGTPGKPGRPGKA 125
Query 67 SAPAAEAAEAEAKKKETE-LLRLAGKPCP 94
AP ++ E + KPCP
Sbjct 126 GAPGLPGNPGRPPQQPCEPITPPPCKPCP 154
Lambda K H
0.310 0.125 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4669349066
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40