bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0192_orf5
Length=354
Score E
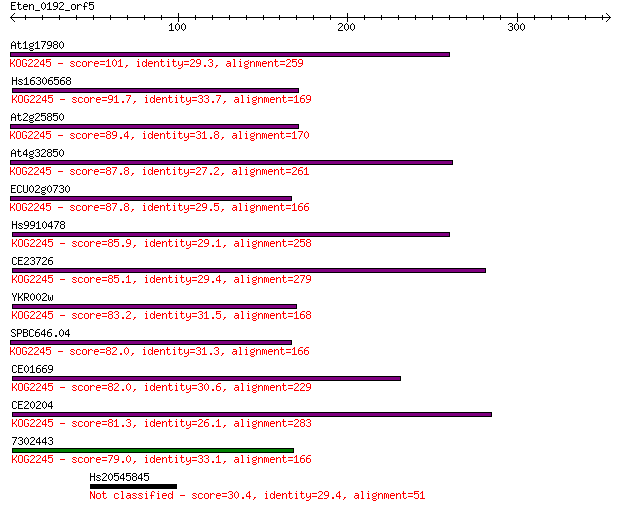
Sequences producing significant alignments: (Bits) Value
At1g17980 101 3e-21
Hs16306568 91.7 2e-18
At2g25850 89.4 1e-17
At4g32850 87.8 3e-17
ECU02g0730 87.8 3e-17
Hs9910478 85.9 1e-16
CE23726 85.1 2e-16
YKR002w 83.2 8e-16
SPBC646.04 82.0 2e-15
CE01669 82.0 2e-15
CE20204 81.3 3e-15
7302443 79.0 1e-14
Hs20545845 30.4 6.1
> At1g17980
Length=714
Score = 101 bits (251), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 76/261 (29%), Positives = 115/261 (44%), Gaps = 51/261 (19%)
Query 1 KLFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAM 60
++F QW W + + LC E G +VW+P I P+DR H++ IITPA+P M
Sbjct 267 RVFYQWNWPNA-IFLCSPDE---------GSLGLQVWDPRINPKDRLHIMPIITPAYPCM 316
Query 61 NSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLR 120
NS++NVS ST R++ E F +E A + A W L
Sbjct 317 NSSYNVSESTLRIMKGE-------FQRGNEICEAMESNKA------------DWDTLF-- 355
Query 121 VLAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSFSSSSSSSSSS 180
P + + ++LQ+ I A + RKW+GWVES++R L +ER F
Sbjct 356 --EPFAFFEAYKNYLQIDISAANVDDLRKWKGWVESRLRQLTLKIER-HFKM-------- 404
Query 181 LNMFIRPLPNVFPFQDPEWPESSSMLIAL--KFASPKAGEAVCDLRPAIAEFVELINGWP 238
+ P P+ FQD P S + L K P A D+R + EF +N +
Sbjct 405 --LHCHPHPH--DFQDTSRPLHCSYFMGLQRKQGVPAAEGEQFDIRRTVEEFKHTVNAY- 459
Query 239 ERAKLAGSIQLRVRHIRKSAL 259
+++ V HI++ +L
Sbjct 460 --TLWIPGMEISVGHIKRRSL 478
> Hs16306568
Length=736
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 57/169 (33%), Positives = 83/169 (49%), Gaps = 33/169 (19%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAMN 61
+F++W W +PV L + E + + VW+P + P DR+HL+ IITPA+P N
Sbjct 274 VFSKWEW-PNPVLLKQPEESNLN---------LPVWDPRVNPSDRYHLMPIITPAYPQQN 323
Query 62 STHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLRV 121
ST+NVS ST+ V+ EE + A DE + W +L
Sbjct 324 STYNVSTSTRTVMVEEFKQG---LAVTDEILQGKS----------------DWSKL---- 360
Query 122 LAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSF 170
L P ++ + H++ L A +E H +W G VESK+R LV LER F
Sbjct 361 LEPPNFFQKYRHYIVLTASASTEENHLEWVGLVESKIRVLVGNLERNEF 409
> At2g25850
Length=414
Score = 89.4 bits (220), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 54/170 (31%), Positives = 81/170 (47%), Gaps = 33/170 (19%)
Query 1 KLFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAM 60
+++TQW W +PV LC I E +F VW+P +DR+HL+ IITPA+P M
Sbjct 277 RVYTQWRW-PNPVMLCAIEEDD---------LSFPVWDPRKNHRDRYHLMPIITPAYPCM 326
Query 61 NSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLR 120
NS++NVS ST RV+ E+ QF + Q W L +
Sbjct 327 NSSYNVSQSTLRVMTEQF-----QFGN--------------TICQEIELNKQHWSSLFQQ 367
Query 121 VLAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSF 170
+ + + ++LQ+ +LA W+GWVES+ R L ++ L F
Sbjct 368 YM----FFEAYKNYLQVDVLAADAEDLLAWKGWVESRFRQLTLKVDSLMF 413
> At4g32850
Length=729
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 71/263 (26%), Positives = 115/263 (43%), Gaps = 49/263 (18%)
Query 1 KLFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAM 60
+++TQW W +PV LC I E E G F VW+ +DR+HL+ IITPA+P M
Sbjct 275 RVYTQWRW-PNPVMLCAIEE---DELG------FPVWDRRKNHRDRYHLMPIITPAYPCM 324
Query 61 NSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLR 120
NS++NVS ST RV+ E+ QF + Q W L +
Sbjct 325 NSSYNVSQSTLRVMTEQF-----QFGN--------------NILQEIELNKQHWSSLFEQ 365
Query 121 VLAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSFSSSSSSSSSS 180
+ + + ++LQ+ I+A W+GWVES+ R L +ER ++
Sbjct 366 YM----FFEAYKNYLQVDIVAADAEDLLAWKGWVESRFRQLTLKIER----------DTN 411
Query 181 LNMFIRPLPNVFPFQDPEWPESSSMLIALKFASPKAGEAV--CDLRPAIAEFVELINGWP 238
+ P PN + ++ + + L+ A G+ D+R + EF + +N +
Sbjct 412 GMLMCHPQPNEYVDTARQFLH-CAFFMGLQRAEGVGGQECQQFDIRGTVDEFRQEVNMY- 469
Query 239 ERAKLAGSIQLRVRHIRKSALGP 261
+ + V H+R+ L P
Sbjct 470 --MFWKPGMDVFVSHVRRRQLPP 490
> ECU02g0730
Length=505
Score = 87.8 bits (216), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 49/166 (29%), Positives = 82/166 (49%), Gaps = 35/166 (21%)
Query 1 KLFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAM 60
+LF+ W W +PV L + + +++ KVW+P +YP D++H + +ITPA+P+M
Sbjct 263 ELFSSWKW-PTPVILRPVVDLNYN---------LKVWDPKVYPSDKYHRMPVITPAYPSM 312
Query 61 NSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLR 120
+THNVS+ST+ VI E A K + P +R R
Sbjct 313 CATHNVSNSTQHVITMEFTRAHK---------------------ILSDPERNDFR----R 347
Query 121 VLAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALE 166
+ D+ S + F+++ ++ E KW+G++ESK+R L E
Sbjct 348 IFELSDFFSRYRLFVEVLAMSSCEEDFPKWEGYLESKIRILASKFE 393
> Hs9910478
Length=636
Score = 85.9 bits (211), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 75/261 (28%), Positives = 119/261 (45%), Gaps = 45/261 (17%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAMN 61
+F++W W +PV L + E+ L VW+P + P DR+HL+ IITPA+P N
Sbjct 275 VFSEWEW-PNPVLLKE-----PEERNLN----LPVWDPRVNPSDRYHLMPIITPAYPQQN 324
Query 62 STHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLRV 121
ST+NVS ST+ V+ EE + A E + A W +L
Sbjct 325 STYNVSISTRMVMIEEFKQG---LAITHEILLSKA----------------EWSKLFE-- 363
Query 122 LAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSFSSSSSSSSSSL 181
AP + + H++ L A +E H +W G VESK+R LV +LE+ F + + + S
Sbjct 364 -AP-SFFQKYKHYIVLLASASTEKQHLEWVGLVESKIRILVGSLEKNEFITLAHVNPQSF 421
Query 182 NMFIRPLPNVFPFQDPEWPESSSM-LIALKFASPKAGEAV-CDLRPAIAEFVELINGWPE 239
P P ++P+ E +M +I L P E + DL I F + +
Sbjct 422 -----PAPK----ENPDMEEFRTMWVIGLGLKKPDNSEILSIDLTYDIQSFTDTVYRQAV 472
Query 240 RAKL-AGSIQLRVRHIRKSAL 259
+K+ +++ H+R+ L
Sbjct 473 NSKMFEMGMKITAMHLRRKEL 493
> CE23726
Length=554
Score = 85.1 bits (209), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 82/300 (27%), Positives = 128/300 (42%), Gaps = 71/300 (23%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAMN 61
+F+ W W +PV L I+ L L VW+P DR+HL+ I+TPAFP N
Sbjct 273 IFSTWKW-PAPVLLDFISSDRMDLAQLNQL----VWDPRRNQSDRYHLMPIVTPAFPQQN 327
Query 62 STHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLRV 121
STHNVS S+ +VI EE++ A D + +W +L
Sbjct 328 STHNVSRSSMKVIQEEMKDA---LITCDNIQNGSC----------------NWMDL---- 364
Query 122 LAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSFSSSSSSSSSSL 181
L I++ S + HF+ L + A++E + G+ ES++R LV LE+
Sbjct 365 LEEINFFSRYKHFISLSMTAETEKDELAFGGFFESRIRQLVLILEK-------------- 410
Query 182 NMFIRPLPNVFP--FQDPEWPESSSMLIALKFASPKAGEAVCDLRPAIAEF--------- 230
N I+ L ++ P F+ + P+ S I L+F DL I +F
Sbjct 411 NQGIK-LAHINPKKFKAAKDPKKSVWFIGLQFDENVKN---LDLTKDIQQFKRNIDYQAK 466
Query 231 ----------VELINGWPERAKLAGSIQLRVRHIRKSALGPEIQQALQEPVSKHRRLEQQ 280
VE + +R+ LA +I ++ K PE ++PVSK R++ +
Sbjct 467 SVKDIEANCAVETDFSYVKRSDLAQTISVKDLKQGKMYKKPES----KDPVSKKRKVSEN 522
> YKR002w
Length=568
Score = 83.2 bits (204), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 53/169 (31%), Positives = 78/169 (46%), Gaps = 34/169 (20%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAMN 61
+ ++W W PV L I + G +VWNP IY QDR H + +ITPA+P+M
Sbjct 262 ILSEWNW-PQPVILKPIED---------GPLQVRVWNPKIYAQDRSHRMPVITPAYPSMC 311
Query 62 STHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLRV 121
+THN++ STK+VI +E + +SW L +
Sbjct 312 ATHNITESTKKVILQEF-------------------VRGVQITNDIFSNKKSWANLFEKN 352
Query 122 LAPIDYLSLFSHFLQLQILAK-SEAVHRKWQGWVESKVRHLVKALERLS 169
D+ + +L++ + S+ H KW G VESKVR LV LE L+
Sbjct 353 ----DFFFRYKFYLEITAYTRGSDEQHLKWSGLVESKVRLLVMKLEVLA 397
> SPBC646.04
Length=566
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/166 (31%), Positives = 77/166 (46%), Gaps = 33/166 (19%)
Query 1 KLFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAM 60
++ QW W + P+ L I + G ++WNP +YP D+ H + IITPA+P+M
Sbjct 260 RILHQWNWPQ-PILLKPIED---------GPLQVRIWNPKLYPSDKAHRMPIITPAYPSM 309
Query 61 NSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLR 120
+THN++ ST+ +I E+ A A A P W L +
Sbjct 310 CATHNITLSTQTIILREM---------------VRAGEIADQIMVKALP----WSALFQK 350
Query 121 VLAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALE 166
D+ + H+L + AK+ KW G VESK+RHLV LE
Sbjct 351 H----DFFHRYKHYLTITAAAKTAEAQLKWAGLVESKLRHLVTRLE 392
> CE01669
Length=554
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 70/233 (30%), Positives = 108/233 (46%), Gaps = 52/233 (22%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFK--VWNPHIYPQDRHHLLKIITPAFPA 59
+F+ W W +PV L +Y + ++ LA VW+P DR+HL+ IITPAFP
Sbjct 268 IFSTWNW-PAPVLL----DYINCDR--TDLAQLNQLVWDPRRNHADRYHLMPIITPAFPQ 320
Query 60 MNSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLL 119
NSTHNVS S+ +VI +E++ A D+ WR+LL
Sbjct 321 QNSTHNVSRSSMKVIQDEMKKA---LIICDKIHEGTL----------------EWRDLL- 360
Query 120 RVLAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSFSSSSSSSSS 179
I++ S + HF+ L++ A+S + G+ ES++R LV+ LE+
Sbjct 361 ---EEINFFSKYHHFIALKLKAESVKEELAFGGFFESRIRQLVQILEK------------ 405
Query 180 SLNMFIRPLPNVFP--FQDPEWPESSSMLIALKFASPKAGEAVCDLRPAIAEF 230
N I+ + + P F+D + P+ S I L+F A DL I +F
Sbjct 406 --NQVIQ-VAQINPRKFKDVKDPKKSIWFIGLEFV---ANVKNVDLTSDIQQF 452
> CE20204
Length=655
Score = 81.3 bits (199), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 74/283 (26%), Positives = 123/283 (43%), Gaps = 50/283 (17%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAMN 61
+F+ WTW PV L E ++ + L VW+P DR H++ IITPAFP N
Sbjct 266 IFSTWTW-PHPVVL---NEMNNDRNDIPTLCEL-VWDPRRKNTDRFHVMPIITPAFPEQN 320
Query 62 STHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLRV 121
STHNV+ ST VI E+ A + E + A +
Sbjct 321 STHNVTRSTATVIKNEICEALEICRDISEGKSKWTA-----------------------L 357
Query 122 LAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALERLSFSSSSSSSSSSL 181
+++ S + HF+ L + A +E + G++ES++R LV++LER + + +
Sbjct 358 FEEVNFFSRYKHFIALIMAAPNEEEELNYGGFLESRIRLLVQSLER---NQDIIIAHNDP 414
Query 182 NMFIRPLPNVFPFQDPEWPESSSMLIALKFASPKAGEAVCDLRPAIAEFVELINGWPERA 241
N +P PN +PE + I L+FA A+ ++L N E
Sbjct 415 NKH-KPSPNAKFDVNPENKRVTVWFIGLEFAEH-------------AKTLDLTN---EIQ 457
Query 242 KLAGSIQLRVRHIRKSALGPEIQQALQEPVSKHRRLEQQLAAA 284
+ +++L+ +++ +GP Q + K L Q ++AA
Sbjct 458 RFKTNVELQASNVK--GIGPNCQVQIDMFYVKRNSLIQVISAA 498
> 7302443
Length=830
Score = 79.0 bits (193), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 55/166 (33%), Positives = 77/166 (46%), Gaps = 33/166 (19%)
Query 2 LFTQWTWNKSPVCLCKITEYSHHEKGLEGLAAFKVWNPHIYPQDRHHLLKIITPAFPAMN 61
+F++W W +PV L H + F+VW+P + DR+HL+ IITPA+P N
Sbjct 283 VFSRWKW-PNPVLL-------KHPDNVN--LRFQVWDPRVNASDRYHLMPIITPAYPQQN 332
Query 62 STHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAAPAAAAATAAAPSLQSWRELLLRV 121
ST NVS STK+VI E + DE W L
Sbjct 333 STFNVSESTKKVILTEFN---RGMNITDEIMLGRIP----------------WERLF--- 370
Query 122 LAPIDYLSLFSHFLQLQILAKSEAVHRKWQGWVESKVRHLVKALER 167
AP + + HF+ L + +++ H +W G VESKVR L+ LER
Sbjct 371 EAP-SFFYRYRHFIVLLVNSQTADDHLEWCGLVESKVRLLIGNLER 415
> Hs20545845
Length=479
Score = 30.4 bits (67), Expect = 6.1, Method: Compositional matrix adjust.
Identities = 15/51 (29%), Positives = 27/51 (52%), Gaps = 0/51 (0%)
Query 48 HLLKIITPAFPAMNSTHNVSHSTKRVIAEELEAAAKQFAAWDEAAAAAAAA 98
HLL+ + P P++ S+ ++ R E+ ++AK AW+E +AA
Sbjct 175 HLLRTLVPQSPSVASSEHMEEENGRSYPEQKVSSAKLTRAWEENNTDGSAA 225
Lambda K H
0.317 0.125 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8559345784
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40