bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0191_orf3
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
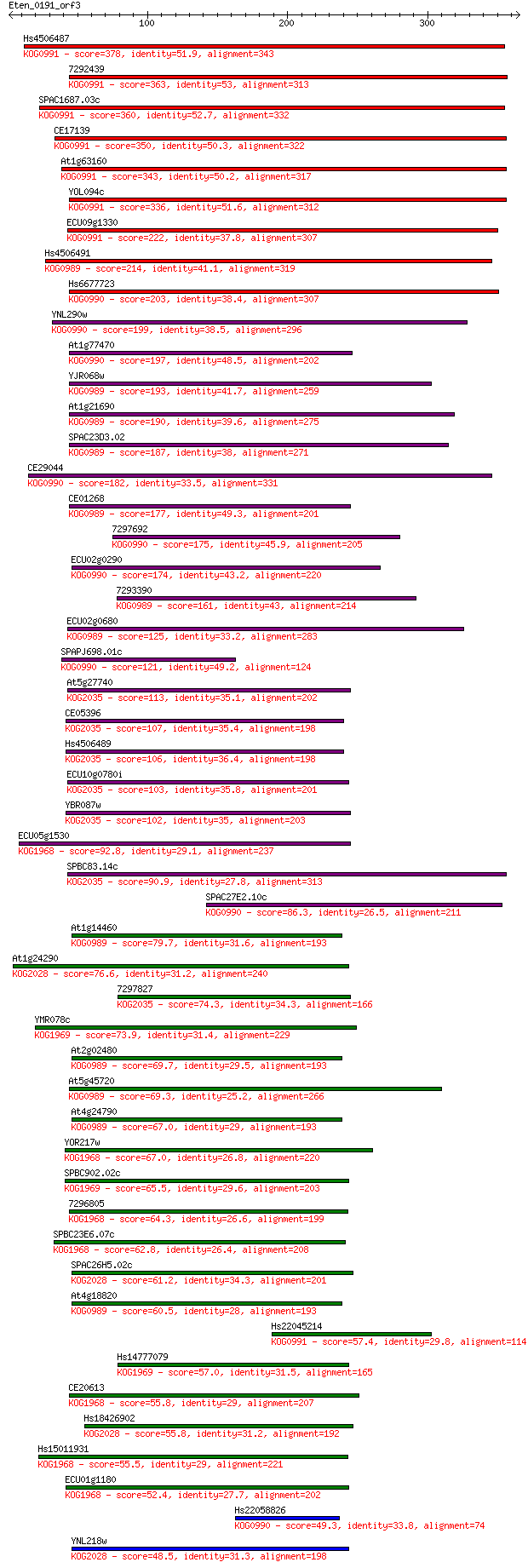
Hs4506487 378 1e-104
7292439 363 4e-100
SPAC1687.03c 360 2e-99
CE17139 350 3e-96
At1g63160 343 3e-94
YOL094c 336 6e-92
ECU09g1330 222 1e-57
Hs4506491 214 2e-55
Hs6677723 203 4e-52
YNL290w 199 1e-50
At1g77470 197 3e-50
YJR068w 193 6e-49
At1g21690 190 4e-48
SPAC23D3.02 187 3e-47
CE29044 182 9e-46
CE01268 177 4e-44
7297692 175 2e-43
ECU02g0290 174 3e-43
7293390 161 2e-39
ECU02g0680 125 2e-28
SPAPJ698.01c 121 3e-27
At5g27740 113 7e-25
CE05396 107 6e-23
Hs4506489 106 8e-23
ECU10g0780i 103 4e-22
YBR087w 102 9e-22
ECU05g1530 92.8 1e-18
SPBC83.14c 90.9 4e-18
SPAC27E2.10c 86.3 9e-17
At1g14460 79.7 8e-15
At1g24290 76.6 7e-14
7297827 74.3 4e-13
YMR078c 73.9 5e-13
At2g02480 69.7 9e-12
At5g45720 69.3 1e-11
At4g24790 67.0 5e-11
YOR217w 67.0 7e-11
SPBC902.02c 65.5 2e-10
7296805 64.3 4e-10
SPBC23E6.07c 62.8 1e-09
SPAC26H5.02c 61.2 3e-09
At4g18820 60.5 6e-09
Hs22045214 57.4 4e-08
Hs14777079 57.0 6e-08
CE20613 55.8 2e-07
Hs18426902 55.8 2e-07
Hs15011931 55.5 2e-07
ECU01g1180 52.4 2e-06
Hs22058826 49.3 1e-05
YNL218w 48.5 2e-05
> Hs4506487
Length=354
Score = 378 bits (971), Expect = 1e-104, Method: Compositional matrix adjust.
Identities = 178/344 (51%), Positives = 249/344 (72%), Gaps = 3/344 (0%)
Query 12 AMNGSSSSSSSSGSSSSSSGSSSSSSSGNESI-WIEKYRPQALDEVVGNDEVLQRLRIIA 70
A+ G + + S + + S + S+G+ + W+EKYRP L+E+VGN++ + RL + A
Sbjct 5 AVCGGAGEVEAQDSDPAPAFSKAPGSAGHYELPWVEKYRPVKLNEIVGNEDTVSRLEVFA 64
Query 71 LEGNMPHLLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFA 130
EGN+P++++AGPPGTGKT+S+LCLARALL + LELNAS++R IDVVR++IK FA
Sbjct 65 REGNVPNIIIAGPPGTGKTTSILCLARALLGPALKDAMLELNASNDRGIDVVRNKIKMFA 124
Query 131 KERRDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSR 190
+++ LP GRHKIIILDE DSMT+ AQQALRR ME++S TTRFALACN+S +IEP+QSR
Sbjct 125 QQKVTLPKGRHKIIILDEADSMTDGAQQALRRTMEIYSKTTRFALACNASDKIIEPIQSR 184
Query 191 CAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFG 250
CA+LR+TKL+DA ++ RL V KE V +TDDG+EAI+F+A GDMR ALNNLQST SGFG
Sbjct 185 CAVLRYTKLTDAQILTRLMNVIEKERVPYTDDGLEAIIFTAQGDMRQALNNLQSTFSGFG 244
Query 251 LVNKENVEKVCDTPPPEMLRRIMQQCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLR 310
+N ENV KVCD P P +++ ++Q CV + A + + LGY+P D++ V +
Sbjct 245 FINSENVFKVCDEPHPLLVKEMIQHCVNANIDEAYKILAHLWHLGYSPEDIIGNIFRVCK 304
Query 311 RADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLCK 354
++ E+L LEF+ +G THM +A G+++ LQM +LA+LC+
Sbjct 305 --TFQMAEYLKLEFIKEIGYTHMKIAEGVNSLLQMAGLLARLCQ 346
> 7292439
Length=331
Score = 363 bits (931), Expect = 4e-100, Method: Compositional matrix adjust.
Identities = 166/313 (53%), Positives = 227/313 (72%), Gaps = 2/313 (0%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSR 103
WIEKYRP E+VGN++ + RL + A +GN P++++AGPPG GKT+++ CLAR LL
Sbjct 18 WIEKYRPVKFKEIVGNEDTVARLSVFATQGNAPNIIIAGPPGVGKTTTIQCLARILLGDS 77
Query 104 WRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRI 163
++ LELNAS+ER IDVVR++IK FA+++ LP GRHKI+ILDE DSMTE AQQALRR
Sbjct 78 YKEAVLELNASNERGIDVVRNKIKMFAQQKVTLPRGRHKIVILDEADSMTEGAQQALRRT 137
Query 164 MELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDG 223
ME++S TTRFALACN+S +IEP+QSRCA+LRFTKLSDA ++ +L EV E + +T+DG
Sbjct 138 MEIYSSTTRFALACNTSEKIIEPIQSRCAMLRFTKLSDAQVLAKLIEVAKWEKLNYTEDG 197
Query 224 IEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIMQQCVAGDWYG 283
+EAIVF+A GDMR LNNLQST GFG + ENV KVCD P P++L ++ C A D +
Sbjct 198 LEAIVFTAQGDMRQGLNNLQSTAQGFGDITAENVFKVCDEPHPKLLEEMIHHCAANDIHK 257
Query 284 AQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTEL 343
A + + LGY+P D++ V +R + + EHL L+F+ +G+THM + G+++ L
Sbjct 258 AYKILAKLWKLGYSPEDIIANIFRVCKRIN--IDEHLKLDFIREIGITHMKIIDGINSLL 315
Query 344 QMEKMLAQLCKVA 356
Q+ +LA+LC A
Sbjct 316 QLTALLAKLCIAA 328
> SPAC1687.03c
Length=342
Score = 360 bits (924), Expect = 2e-99, Method: Compositional matrix adjust.
Identities = 175/332 (52%), Positives = 238/332 (71%), Gaps = 1/332 (0%)
Query 23 SGSSSSSSGSSSSSSSGNESIWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAG 82
S + SSS ++S E W+EKYRP LD++VGN+E + RL++IA EGNMPHL+++G
Sbjct 2 SNAVSSSVFGEKNNSVAYELPWVEKYRPIVLDDIVGNEETIDRLKVIAKEGNMPHLVISG 61
Query 83 PPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHK 142
PG GKT+S+LCLA ALL ++ LELNASDER IDVVR+RIKAFA+++ LP GRHK
Sbjct 62 MPGIGKTTSILCLAHALLGPAYKEGVLELNASDERGIDVVRNRIKAFAQKKVILPPGRHK 121
Query 143 IIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDA 202
IIILDE DSMT AQQALRR ME++S+TTRFALACN S+ +IEP+QSRCAILR+++L+D
Sbjct 122 IIILDEADSMTAGAQQALRRTMEIYSNTTRFALACNQSNKIIEPIQSRCAILRYSRLTDQ 181
Query 203 HLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCD 262
++QRL +C E V +TDDG+ A++ +A+GDMR A+NNLQSTV+GFGLVN ENV +V D
Sbjct 182 QVLQRLLNICKAEKVNYTDDGLAALIMTAEGDMRQAVNNLQSTVAGFGLVNGENVFRVAD 241
Query 263 TPPPEMLRRIMQQCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLL 322
P P + ++ C +G+ A + I LG++ +D+VT V++ DS + E L
Sbjct 242 QPSPVAIHAMLTACQSGNIDVALEKLQGIWDLGFSAVDIVTNMFRVVKTMDS-IPEFSRL 300
Query 323 EFLGIVGMTHMTMAGGLSTELQMEKMLAQLCK 354
E L +G THM + G+ T LQ+ ++ +L K
Sbjct 301 EMLKEIGQTHMIILEGVQTLLQLSGLVCRLAK 332
> CE17139
Length=334
Score = 350 bits (898), Expect = 3e-96, Method: Compositional matrix adjust.
Identities = 162/324 (50%), Positives = 231/324 (71%), Gaps = 2/324 (0%)
Query 34 SSSSSGNESIWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVL 93
S S + W+EKYRP+ L ++VGN+ +++RL++I EGN+P+++L+GPPG GKT+SV
Sbjct 2 SKSEKQQLAPWVEKYRPKVLADIVGNENIVERLKVIGHEGNVPNIVLSGPPGCGKTTSVW 61
Query 94 CLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMT 153
LAR LL + + LELNASDER IDVVR RIK FA+ + LP GRHKIIILDE DSMT
Sbjct 62 ALARELLGDKVKEAVLELNASDERGIDVVRHRIKTFAQTKVTLPEGRHKIIILDEADSMT 121
Query 154 EAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCA 213
+ AQQALRR ME+++ TTRFALACN S +IEP+QSRCA+LR+TKLS L+ R++EV
Sbjct 122 DGAQQALRRTMEMYTKTTRFALACNQSEKIIEPIQSRCALLRYTKLSPVQLLTRVKEVAK 181
Query 214 KENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIM 273
E V + D G+EAI+F+A GDMR ALNNLQ+TV+ + LVNKENV KVCD P P+++ +++
Sbjct 182 AEKVNYDDGGLEAILFTAQGDMRQALNNLQATVNAYELVNKENVLKVCDEPHPDLMIKML 241
Query 274 QQCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRAD--SELQEHLLLEFLGIVGMT 331
C ++ A + LG++ D+V+T V++ + + E L +E++ + M
Sbjct 242 HYCTDRKFFEASKIIHEFHRLGFSSDDIVSTLFRVVKTVELSKNVSEQLRMEYIRQIAMC 301
Query 332 HMTMAGGLSTELQMEKMLAQLCKV 355
HM + GL+++LQ+ +++A LC+V
Sbjct 302 HMRIVQGLTSKLQLSRLIADLCRV 325
> At1g63160
Length=333
Score = 343 bits (880), Expect = 3e-94, Method: Compositional matrix adjust.
Identities = 159/317 (50%), Positives = 230/317 (72%), Gaps = 2/317 (0%)
Query 39 GNESIWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARA 98
G W+EKYRP + ++VGN++ + RL++IA +GNMP+L+L+GPPGTGKT+S+L LA
Sbjct 12 GYNEPWVEKYRPSKVVDIVGNEDAVSRLQVIARDGNMPNLILSGPPGTGKTTSILALAHE 71
Query 99 LLQSRWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQ 158
LL + ++ LELNASD+R IDVVR++IK FA+++ LP GRHK++ILDE DSMT AQQ
Sbjct 72 LLGTNYKEAVLELNASDDRGIDVVRNKIKMFAQKKVTLPPGRHKVVILDEADSMTSGAQQ 131
Query 159 ALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVT 218
ALRR +E++S++TRFALACN+S+ +IEP+QSRCA++RF++LSD ++ RL V A E V
Sbjct 132 ALRRTIEIYSNSTRFALACNTSAKIIEPIQSRCALVRFSRLSDQQILGRLLVVVAAEKVP 191
Query 219 FTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIMQQCVA 278
+ +G+EAI+F+ADGDMR ALNNLQ+T SGF VN+ENV KVCD P P ++ I++ +
Sbjct 192 YVPEGLEAIIFTADGDMRQALNNLQATFSGFSFVNQENVFKVCDQPHPLHVKNIVRNVLE 251
Query 279 GDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGG 338
+ A + + LGY+P D++TT +++ D + E+L LEF+ G HM + G
Sbjct 252 SKFDIACDGLKQLYDLGYSPTDIITTLFRIIKNYD--MAEYLKLEFMKETGFAHMRICDG 309
Query 339 LSTELQMEKMLAQLCKV 355
+ + LQ+ +LA+L V
Sbjct 310 VGSYLQLCGLLAKLSIV 326
> YOL094c
Length=323
Score = 336 bits (861), Expect = 6e-92, Method: Compositional matrix adjust.
Identities = 161/314 (51%), Positives = 234/314 (74%), Gaps = 7/314 (2%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSR 103
W+EKYRPQ L ++VGN E + RL+ IA +GNMPH++++G PG GKT+SV CLA LL
Sbjct 11 WVEKYRPQVLSDIVGNKETIDRLQQIAKDGNMPHMIISGMPGIGKTTSVHCLAHELLGRS 70
Query 104 WRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRI 163
+ LELNASD+R IDVVR++IK FA+++ LP G+HKI+ILDE DSMT AQQALRR
Sbjct 71 YADGVLELNASDDRGIDVVRNQIKHFAQKKLHLPPGKHKIVILDEADSMTAGAQQALRRT 130
Query 164 MELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDG 223
MEL+S++TRFA ACN S+ +IEPLQSRCAILR++KLSD +++RL ++ E+V +T+DG
Sbjct 131 MELYSNSTRFAFACNQSNKIIEPLQSRCAILRYSKLSDEDVLKRLLQIIKLEDVKYTNDG 190
Query 224 IEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIMQQCVAGDWYG 283
+EAI+F+A+GDMR A+NNLQSTV+G GLVN +NV K+ D+P P ++++++ +A +
Sbjct 191 LEAIIFTAEGDMRQAINNLQSTVAGHGLVNADNVFKIVDSPHPLIVKKML---LASNLED 247
Query 284 AQSVARVIL-SLGYTPLDVVTT-FRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLST 341
+ + R L GY+ +D+VTT FR + + ++++E + LE + +G+THM + G+ T
Sbjct 248 SIQILRTDLWKKGYSSIDIVTTSFR--VTKNLAQVKESVRLEMIKEIGLTHMRILEGVGT 305
Query 342 ELQMEKMLAQLCKV 355
LQ+ MLA++ K+
Sbjct 306 YLQLASMLAKIHKL 319
> ECU09g1330
Length=309
Score = 222 bits (565), Expect = 1e-57, Method: Compositional matrix adjust.
Identities = 116/308 (37%), Positives = 190/308 (61%), Gaps = 13/308 (4%)
Query 43 IWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQS 102
+ + KY+P + ++VGN+ ++ + ++ +MPHLL GPPGTGKT+ LAR LL +
Sbjct 3 LLVNKYQPSEIQDIVGNEATMELVSLMIESRDMPHLLFTGPPGTGKTTCAKILARRLLGN 62
Query 103 RWRSCCLELNASDERSIDVVRDRIKAFAKER-RDLPLGRHKIIILDEVDSMTEAAQQALR 161
+ LELNASDER ID VR IK+FA+ R +D KIIILDE DSMT AQQA+R
Sbjct 63 K--EGLLELNASDERGIDTVRTTIKSFAQRRVKDCEF---KIIILDEADSMTTTAQQAMR 117
Query 162 RIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTD 221
R+ME+HS RF L CN + + EP+QSRCAILRF ++ + +++RL+E+ E + T
Sbjct 118 RVMEIHSSECRFILICNVFTKIFEPIQSRCAILRFDRIEQSVILKRLKEISEGEGIRITA 177
Query 222 DGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIMQQCVAGDW 281
+ ++ +V +DGDMR +LN LQ+ ++ G V+++ + K+ P P+ + +++Q+ + +
Sbjct 178 EALDLVVELSDGDMRQSLNILQACINSPGTVDQDYIIKIIGLPSPKRIEKVLQRLLKREV 237
Query 282 YGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLST 341
A + I + PLD++ +F RA ++ + E L ++G+ ++ ++ G+++
Sbjct 238 EEALEMFDEIWEEKFDPLDLINSF----FRAAKNMESY---ELLKVIGLANLRISEGVNS 290
Query 342 ELQMEKML 349
LQ M
Sbjct 291 RLQFYGMF 298
> Hs4506491
Length=363
Score = 214 bits (545), Expect = 2e-55, Method: Compositional matrix adjust.
Identities = 131/330 (39%), Positives = 190/330 (57%), Gaps = 15/330 (4%)
Query 27 SSSSGSSSSSSSGNESIWIEKYRPQALDEVVGNDEVLQRLRIIALEG-NMPHLLLAGPPG 85
++S+GSS + W+EKYRP+ +DEV +EV+ L+ +LEG ++P+LL GPPG
Sbjct 23 AASAGSSGENKKAKPVPWVEKYRPKCVDEVAFQEEVVAVLKK-SLEGADLPNLLFYGPPG 81
Query 86 TGKTSSVLCLARALLQSR-WRSCCLELNASDERSIDVVRDRIKAFAK-----ERRD-LPL 138
TGKTS++L AR L +R LELNASDER I VVR+++K FA+ R D P
Sbjct 82 TGKTSTILAAARELFGPELFRLRVLELNASDERGIQVVREKVKNFAQLTVSGSRSDGKPC 141
Query 139 GRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTK 198
KI+ILDE DSMT AAQ ALRR ME S TTRF L CN S +IEPL SRC+ RF
Sbjct 142 PPFKIVILDEADSMTSAAQAALRRTMEKESKTTRFCLICNYVSRIIEPLTSRCSKFRFKP 201
Query 199 LSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQST--VSGFGLVNKEN 256
LSD QRL ++ KENV +D+GI +V ++GD+R A+ LQS ++G + ++
Sbjct 202 LSDKIQQQRLLDIAKKENVKISDEGIAYLVKVSEGDLRKAITFLQSATRLTGGKEITEKV 261
Query 257 VEKVCDTPPPEMLRRIMQQCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRAD-SE 315
+ + P E + + C +G + ++V + ++ G+ +V V+ + S+
Sbjct 262 ITDIAGVIPAEKIDGVFAACQSGSFDKLEAVVKDLIDEGHAATQLVNQLHDVVVENNLSD 321
Query 316 LQEHLLLEFLGIVGMTHMTMAGGLSTELQM 345
Q+ ++ E L V +A G LQ+
Sbjct 322 KQKSIITEKLAEV---DKCLADGADEHLQL 348
> Hs6677723
Length=340
Score = 203 bits (517), Expect = 4e-52, Method: Compositional matrix adjust.
Identities = 118/311 (37%), Positives = 180/311 (57%), Gaps = 9/311 (2%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSR 103
W+EKYRPQ L++++ + ++L ++ E +PHLLL GPPGTGKTS++L A+ L + +
Sbjct 22 WVEKYRPQTLNDLISHQDILSTIQKFINEDRLPHLLLYGPPGTGKTSTILACAKQLYKDK 81
Query 104 -WRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRR 162
+ S LELNASD+R ID++R I +FA R G K++ILDE D+MT+ AQ ALRR
Sbjct 82 EFGSMVLELNASDDRGIDIIRGPILSFASTRTIFKKG-FKLVILDEADAMTQDAQNALRR 140
Query 163 IMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDD 222
++E ++ TRF L CN S +I LQSRC RF L+ +V RL V +E V ++D
Sbjct 141 VIEKFTENTRFCLICNYLSKIIPALQSRCTRFRFGPLTPELMVPRLEHVVEEEKVDISED 200
Query 223 GIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIMQQCVAGDWY 282
G++A+V + GDMR ALN LQST FG V +E V P + I+ + D+
Sbjct 201 GMKALVTLSSGDMRRALNILQSTNMAFGKVTEETVYTCTGHPLKSDIANILDWMLNQDFT 260
Query 283 GA-QSVARVILSLGYTPLDVVTTFRGVLRRAD--SELQEHLLLEFLGIVGMTHMTMAGGL 339
A +++ + G D++T + R D S ++ HLL + I ++ G
Sbjct 261 TAYRNITELKTLKGLALHDILTEIHLFVHRVDFPSSVRIHLLTKMADI----EYRLSVGT 316
Query 340 STELQMEKMLA 350
+ ++Q+ ++A
Sbjct 317 NEKIQLSSLIA 327
> YNL290w
Length=340
Score = 199 bits (505), Expect = 1e-50, Method: Compositional matrix adjust.
Identities = 114/308 (37%), Positives = 174/308 (56%), Gaps = 15/308 (4%)
Query 32 SSSSSSSGNESI-WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTS 90
S+S+ E++ W+EKYRP+ LDEV G +EV+ +R EG +PHLL GPPGTGKTS
Sbjct 2 STSTEKRSKENLPWVEKYRPETLDEVYGQNEVITTVRKFVDEGKLPHLLFYGPPGTGKTS 61
Query 91 SVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVD 150
+++ LAR + + + LELNASD+R IDVVR++IK FA R+ G K+IILDE D
Sbjct 62 TIVALAREIYGKNYSNMVLELNASDDRGIDVVRNQIKDFASTRQIFSKG-FKLIILDEAD 120
Query 151 SMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLRE 210
+MT AAQ ALRR++E ++ TRF + N + + L SRC RF L + +R+
Sbjct 121 AMTNAAQNALRRVIERYTKNTRFCVLANYAHKLTPALLSRCTRFRFQPLPQEAIERRIAN 180
Query 211 VCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVE-----KVCDTPP 265
V E + + + +A++ ++GDMR LN LQS + +++ + + C P
Sbjct 181 VLVHEKLKLSPNAEKALIELSNGDMRRVLNVLQSCKATLDNPDEDEISDDVIYECCGAPR 240
Query 266 PEMLRRIMQQCVAGDWYGAQ-SVARVILSLGYTPLDVVTTFRGVLRRADSELQE-----H 319
P L+ +++ + DW A ++ +V + G +D++ +L D ELQ H
Sbjct 241 PSDLKAVLKSILEDDWGTAHYTLNKVRSAKGLALIDLIEGIVKILE--DYELQNEETRVH 298
Query 320 LLLEFLGI 327
LL + I
Sbjct 299 LLTKLADI 306
> At1g77470
Length=369
Score = 197 bits (501), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 98/204 (48%), Positives = 139/204 (68%), Gaps = 3/204 (1%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSR 103
W+EKYRPQ+LD+V + +++ + + E +PHLLL GPPGTGKTS++L +AR L +
Sbjct 41 WVEKYRPQSLDDVAAHRDIIDTIDRLTNENKLPHLLLYGPPGTGKTSTILAVARKLYGPK 100
Query 104 WRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRH--KIIILDEVDSMTEAAQQALR 161
+R+ LELNASD+R IDVVR +I+ FA + LG+ K+++LDE D+MT+ AQ ALR
Sbjct 101 YRNMILELNASDDRGIDVVRQQIQDFAST-QSFSLGKSSVKLVLLDEADAMTKDAQFALR 159
Query 162 RIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTD 221
R++E ++ +TRFAL N + +I LQSRC RF L H+ QRL+ V E + +D
Sbjct 160 RVIEKYTKSTRFALIGNHVNKIIPALQSRCTRFRFAPLDGVHMSQRLKHVIEAERLVVSD 219
Query 222 DGIEAIVFSADGDMRSALNNLQST 245
G+ A+V ++GDMR ALN LQST
Sbjct 220 CGLAALVRLSNGDMRKALNILQST 243
> YJR068w
Length=353
Score = 193 bits (490), Expect = 6e-49, Method: Compositional matrix adjust.
Identities = 108/275 (39%), Positives = 154/275 (56%), Gaps = 16/275 (5%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALL-QS 102
W+EKYRP+ LDEV D + L+ N+PH+L GPPGTGKTS++L L + L
Sbjct 27 WVEKYRPKNLDEVTAQDHAVTVLKKTLKSANLPHMLFYGPPGTGKTSTILALTKELYGPD 86
Query 103 RWRSCCLELNASDERSIDVVRDRIKAFAK------ERRDL---PLGRHKIIILDEVDSMT 153
+S LELNASDER I +VR+++K FA+ + DL P +KIIILDE DSMT
Sbjct 87 LMKSRILELNASDERGISIVREKVKNFARLTVSKPSKHDLENYPCPPYKIIILDEADSMT 146
Query 154 EAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCA 213
AQ ALRR ME +S TRF L CN + +I+PL SRC+ RF L ++ + RLR +
Sbjct 147 ADAQSALRRTMETYSGVTRFCLICNYVTRIIDPLASRCSKFRFKALDASNAIDRLRFISE 206
Query 214 KENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGL------VNKENVEKVCDTPPPE 267
+ENV D +E I+ + GD+R + LQS G + VE++ P +
Sbjct 207 QENVKCDDGVLERILDISAGDLRRGITLLQSASKGAQYLGDGKNITSTQVEELAGVVPHD 266
Query 268 MLRRIMQQCVAGDWYGAQSVARVILSLGYTPLDVV 302
+L I+++ +GD+ + + G++ VV
Sbjct 267 ILIEIVEKVKSGDFDEIKKYVNTFMKSGWSAASVV 301
> At1g21690
Length=336
Score = 190 bits (483), Expect = 4e-48, Method: Compositional matrix adjust.
Identities = 109/297 (36%), Positives = 158/297 (53%), Gaps = 22/297 (7%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSR 103
W+EKYRP+ + +V +EV++ L + PH+L GPPGTGKT++ L +A L
Sbjct 11 WVEKYRPKQVKDVAHQEEVVRVLTNTLQTADCPHMLFYGPPGTGKTTTALAIAHQLFGPE 70
Query 104 -WRSCCLELNASDERSIDVVRDRIKAFA-------KERRDLPLGRHKIIILDEVDSMTEA 155
++S LELNASD+R I+VVR +IK FA + P KIIILDE DSMTE
Sbjct 71 LYKSRVLELNASDDRGINVVRTKIKDFAAVAVGSNHRQSGYPCPSFKIIILDEADSMTED 130
Query 156 AQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKE 215
AQ ALRR ME +S TRF CN S +IEPL SRCA RF LS+ + R+ +C +E
Sbjct 131 AQNALRRTMETYSKVTRFFFICNYISRIIEPLASRCAKFRFKPLSEEVMSNRILHICNEE 190
Query 216 NVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFG-------LVNKENVE-------KVC 261
++ + + + + GD+R A+ LQS FG L+N V C
Sbjct 191 GLSLDGEALSTLSSISQGDLRRAITYLQSATRLFGSTITSTDLLNVSGVRMKLTQYLHKC 250
Query 262 DTPPPEMLRRIMQQCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQE 318
P E++ ++ C +GD+ A I++ GY ++ ++ ADS++ +
Sbjct 251 MVVPLEVVNKLFTACKSGDFDIANKEVDNIVAEGYPASQIINQLFDIVAEADSDITD 307
> SPAC23D3.02
Length=340
Score = 187 bits (476), Expect = 3e-47, Method: Compositional matrix adjust.
Identities = 103/279 (36%), Positives = 164/279 (58%), Gaps = 8/279 (2%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSR 103
W+E YRP+ LD+V + +Q L+ L N+PH+L G PGTGKTS++L L+R L +
Sbjct 21 WVELYRPKTLDQVSSQESTVQVLKKTLLSNNLPHMLFYGSPGTGKTSTILALSRELFGPQ 80
Query 104 -WRSCCLELNASDERSIDVVRDRIKAFAK----ERRD-LPLGRHKIIILDEVDSMTEAAQ 157
+S LELNASDER I ++R+++K+FAK + D P KIIILDE DSMT+ AQ
Sbjct 81 LMKSRVLELNASDERGISIIREKVKSFAKTTVTNKVDGYPCPPFKIIILDEADSMTQDAQ 140
Query 158 QALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENV 217
ALRR ME ++ TRF L CN + +I+PL SRC+ RF L + ++V+RL + A + V
Sbjct 141 AALRRTMESYARITRFCLICNYMTRIIDPLSSRCSKYRFKPLDNENMVKRLEFIAADQAV 200
Query 218 TFTDDGIEAIVFSADGDMRSALNNLQSTVSGF--GLVNKENVEKVCDTPPPEMLRRIMQQ 275
+ + A+V + GDMR A+ LQS + + +VE++ P ++R ++
Sbjct 201 SMEPGVVNALVECSGGDMRKAITFLQSAANLHQGTPITISSVEELAGAVPYNIIRSLLDT 260
Query 276 CVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRADS 314
+ ++++R + + GY+ +++ VL + ++
Sbjct 261 AYTKNVSNIETLSRDVAAEGYSTGIILSQLHDVLLKEET 299
> CE29044
Length=388
Score = 182 bits (463), Expect = 9e-46, Method: Compositional matrix adjust.
Identities = 111/364 (30%), Positives = 182/364 (50%), Gaps = 38/364 (10%)
Query 15 GSSSSSSSSGSSSSSSGSSSSSSSGNESIWIEKYRPQALDEVVGNDEVLQRLRIIALEGN 74
GS + S S + + ++++ + W+EKYRP LDE+V ++++++ L
Sbjct 2 GSRTGSREEHSLQFLNLTKMTTTTASNLPWVEKYRPSKLDELVAHEQIVKTLTKFIENRT 61
Query 75 MPHLLLAGPPGTGKTSSVLCLARALLQ-SRWRSCCLELNASDERSIDVVRDRIKAFAKER 133
+PHLL GPPGTGKT++VL AR + ++ S LELNASDER IDVVR+ I FA+ +
Sbjct 62 LPHLLFYGPPGTGKTTTVLAAARQMYSPTKMASMVLELNASDERGIDVVRNTIVNFAQTK 121
Query 134 R-----------DLPLGRHKIIILDEVDSMTEAAQQALRR-------------------- 162
+P K++ILDE D+MT+ AQ ALRR
Sbjct 122 GLQAFSTSSNTGTVPF---KLVILDEADAMTKDAQNALRRLDKTDYEQNIIIHNVNIYFR 178
Query 163 IMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDD 222
++E ++D RF + CN +S++ +QSRC RF L +V RL + E + T D
Sbjct 179 VIEKYTDNVRFCIICNYLASIVPAIQSRCTRFRFAPLDQKLIVPRLEYIVETEQLKMTPD 238
Query 223 GIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEMLRRIMQQCVAGDWY 282
G +A++ + GDMR+ +N LQST F V++ V + P P+ ++ +++ +
Sbjct 239 GKDALLIVSKGDMRTVINTLQSTAMSFDTVSENTVYQCIGQPTPKEMKEVVKTLLNDPSK 298
Query 283 GA-QSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHLLLEFLGIVGMTHMTMAGGLST 341
++ + GY DV+T + D + + + + +G ++ G S
Sbjct 299 KCMNTIQTKLFENGYALQDVITHLHDFVFTLD--IPDEAMSAIITGLGEVEENLSTGCSN 356
Query 342 ELQM 345
E Q+
Sbjct 357 ETQL 360
> CE01268
Length=334
Score = 177 bits (448), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 99/209 (47%), Positives = 134/209 (64%), Gaps = 11/209 (5%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLRIIALEG-NMPHLLLAGPPGTGKTSSVLCLARALL-Q 101
W EKYRP+ LD++ DEV+ L+ AL+G ++PHLL GPPGTGKTS+ L R L +
Sbjct 17 WTEKYRPKTLDDIAYQDEVVTMLKG-ALQGRDLPHLLFYGPPGTGKTSAALAFCRQLFPK 75
Query 102 SRWRSCCLELNASDERSIDVVRDRIKAFAK------ERRDLPLGRHKIIILDEVDSMTEA 155
+ + L+LNASDER I VVR +I++F+K R D+ + KIIILDEVD+MT
Sbjct 76 NIFHDRVLDLNASDERGIAVVRQKIQSFSKSSLGHSHREDVL--KLKIIILDEVDAMTRE 133
Query 156 AQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKE 215
AQ A+RR++E S TTRF L CN S +I P+ SRCA RF L VQRLR +C E
Sbjct 134 AQAAMRRVIEDFSKTTRFILICNYVSRLIPPVVSRCAKFRFKSLPAEIQVQRLRTICDAE 193
Query 216 NVTFTDDGIEAIVFSADGDMRSALNNLQS 244
+DD ++ ++ ++GD+R A+ LQS
Sbjct 194 GTPMSDDELKQVMEYSEGDLRRAVCTLQS 222
> 7297692
Length=297
Score = 175 bits (443), Expect = 2e-43, Method: Compositional matrix adjust.
Identities = 94/206 (45%), Positives = 127/206 (61%), Gaps = 2/206 (0%)
Query 75 MPHLLLAGPPGTGKTSSVLCLARALLQ-SRWRSCCLELNASDERSIDVVRDRIKAFAKER 133
+PHLL GPPGTGKTS++L AR L +++S LELNASD+R I +VR +I FA R
Sbjct 9 LPHLLFYGPPGTGKTSTILACARQLYSPQQFKSMVLELNASDDRGIGIVRGQILNFASTR 68
Query 134 RDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAI 193
+ K+IILDE D+MT AQ ALRRI+E ++D RF + CN S +I LQSRC
Sbjct 69 -TIFCDTFKLIILDEADAMTNDAQNALRRIIEKYTDNVRFCVICNYLSKIIPALQSRCTR 127
Query 194 LRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVN 253
RF LS ++ RL ++ E V T+DG A++ A GDMR LN LQSTV F VN
Sbjct 128 FRFAPLSQDQMMPRLEKIIEAEAVQITEDGKRALLTLAKGDMRKVLNVLQSTVMAFDTVN 187
Query 254 KENVEKVCDTPPPEMLRRIMQQCVAG 279
++NV P + + +I++ ++G
Sbjct 188 EDNVYMCVGYPLRQDIEQILKALLSG 213
> ECU02g0290
Length=305
Score = 174 bits (441), Expect = 3e-43, Method: Compositional matrix adjust.
Identities = 95/220 (43%), Positives = 132/220 (60%), Gaps = 10/220 (4%)
Query 46 EKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQSRWR 105
EKYRP +L EVVGN EV+ L+ I+ G +P++L GPPGTGKT+S+ RA+ + R
Sbjct 7 EKYRPGSLLEVVGNREVVSSLQSISSAGRIPNMLFYGPPGTGKTTSI----RAIANNLPR 62
Query 106 SCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRIME 165
+ LELNASDER I VR+ IK FA K++ILDE D M+ AQ ALRRI+E
Sbjct 63 TSVLELNASDERGIATVRETIKEFASTYSKT----MKLVILDEADMMSRDAQNALRRIIE 118
Query 166 LHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIE 225
S TRF L N +I P+ SRC RF + D R+ E+C +E++ +T +GI+
Sbjct 119 DFSANTRFCLIANHLRKIISPILSRCTKFRFGPIGDTE--NRIEEICKRESIKYTPEGIK 176
Query 226 AIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPP 265
A+ + GDMR A+N++Q + G V++ NV + P
Sbjct 177 AVSRISMGDMRKAVNDIQGISASLGQVDESNVRRFNGIAP 216
> 7293390
Length=288
Score = 161 bits (407), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 92/223 (41%), Positives = 129/223 (57%), Gaps = 9/223 (4%)
Query 78 LLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFAK-----E 132
+LL GPPGTGKTS++L +R + ++ LELNASDER I+VVR +IK F++
Sbjct 1 MLLYGPPGTGKTSTILAASRQIFGDMFKDRILELNASDERGINVVRTKIKNFSQLSASSV 60
Query 133 RRD-LPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRC 191
R D P KIIILDE DSMT AAQ ALRR ME S +TRF L CN S +I P+ SRC
Sbjct 61 RPDGKPCPPFKIIILDEADSMTHAAQSALRRTMEKESRSTRFCLICNYVSRIIVPITSRC 120
Query 192 AILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFG- 250
+ RF L + ++ RL+ +C E V DD ++IV + GD+R A+ LQS G
Sbjct 121 SKFRFKALGEDKVIDRLKYICEMEGVKIEDDAYKSIVKISGGDLRRAITTLQSCYRLKGP 180
Query 251 --LVNKENVEKVCDTPPPEMLRRIMQQCVAGDWYGAQSVARVI 291
++N ++ ++ P L ++ C +G++ + R I
Sbjct 181 EHIINTADLFEMSGVIPEYYLEDYLEVCRSGNYERLEQFVREI 223
> ECU02g0680
Length=283
Score = 125 bits (314), Expect = 2e-28, Method: Compositional matrix adjust.
Identities = 94/292 (32%), Positives = 141/292 (48%), Gaps = 45/292 (15%)
Query 43 IWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQS 102
+W EKYRP++ D G + + LR + G+ P+LLL GPPGTGKT+ LA
Sbjct 2 LWTEKYRPKSTDAFEGPEHLKNILRNSSGRGH-PNLLLYGPPGTGKTTFAHLLA------ 54
Query 103 RWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRR 162
S LELNASDER I V+R++IK +A LG+ K +ILDE +++T AQ LRR
Sbjct 55 ---SQKLELNASDERGISVIREKIKVYAS-----TLGKDKTVILDECENLTSDAQHCLRR 106
Query 163 IMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDD 222
++E S TRF N S +I PL+SR ++FT + L + +KE + + +
Sbjct 107 VIE-DSVNTRFIFITNYPSKIIGPLRSRLVSVKFTPTES----KILENIGSKEGLGYDKE 161
Query 223 GIEAIVFSADGDMRSALNNLQSTV--------SGFGLVNKENVEKVCDTPPPEMLRRIMQ 274
I+ D+R A+N LQ G V ++ V+ + P ++ +
Sbjct 162 LYHRILKLCGNDLRRAINVLQGVAPLGSFCIEEAVGAVPQKTVDMFWEVDRPRVVEYV-- 219
Query 275 QCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVLRRADSELQE-HLLLEFL 325
R L GY+ L ++ G + +D++ E HL L L
Sbjct 220 --------------REFLREGYSALQLIHQLAGSSKGSDAQSAELHLALSLL 257
> SPAPJ698.01c
Length=142
Score = 121 bits (303), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 61/126 (48%), Positives = 84/126 (66%), Gaps = 3/126 (2%)
Query 39 GNESI--WIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLA 96
G+ES W+EKYRP L++VV + +++ L +PH+L GPPGTGKTS++L A
Sbjct 18 GSESTLPWVEKYRPANLEDVVSHKDIISTLEKFISSNRVPHMLFYGPPGTGKTSTILACA 77
Query 97 RALLQSRWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAA 156
R + +R+ +ELNASD+R ID VR++IK FA R+ + K+IILDE D+MT AA
Sbjct 78 RKIYGPNYRNQLMELNASDDRGIDAVREQIKNFASTRQ-IFASTFKMIILDEADAMTLAA 136
Query 157 QQALRR 162
Q ALRR
Sbjct 137 QNALRR 142
> At5g27740
Length=354
Score = 113 bits (282), Expect = 7e-25, Method: Compositional matrix adjust.
Identities = 71/233 (30%), Positives = 126/233 (54%), Gaps = 31/233 (13%)
Query 43 IWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARAL--- 99
+W++KYRP++LD+V+ ++++ Q+L+ + E + PHLL GP G+GK + ++ L + +
Sbjct 2 LWVDKYRPKSLDKVIVHEDIAQKLKKLVSEQDCPHLLFYGPSGSGKKTLIMALLKQIYGA 61
Query 100 ------LQSR-WR-----------------SCCLELNASDERSID--VVRDRIKAFAKER 133
+++R W+ + +EL SD D +V++ IK AK R
Sbjct 62 SAEKVKVENRAWKVDAGSRTIDLELTTLSSTNHVELTPSDAGFQDRYIVQEIIKEMAKNR 121
Query 134 RDLPLGR--HKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRC 191
G+ +K+++L+EVD ++ AQ +LRR ME +S + R L CNSSS V E ++SRC
Sbjct 122 PIDTKGKKGYKVLVLNEVDKLSREAQHSLRRTMEKYSSSCRLILCCNSSSKVTEAIKSRC 181
Query 192 AILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQS 244
+R S +V+ L V KE++ I ++ +R A+ +L++
Sbjct 182 LNVRINAPSQEEIVKVLEFVAKKESLQLPQGFAARIAEKSNRSLRRAILSLET 234
> CE05396
Length=354
Score = 107 bits (266), Expect = 6e-23, Method: Compositional matrix adjust.
Identities = 70/229 (30%), Positives = 118/229 (51%), Gaps = 32/229 (13%)
Query 42 SIWIEKYRPQAL---DEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARA 98
++W++KYRP+ L D V + E L+ ++ + MPHLL GP G GK + + CL R
Sbjct 2 ALWVDKYRPKDLLGKDGVDYHIEQANHLKFLSADC-MPHLLFCGPSGAGKKTRIKCLLRE 60
Query 99 L------------------------LQSRWRSCCLELNASDERSID--VVRDRIKAFAKE 132
L +Q+ + +E+ SD D VV+D +K A+
Sbjct 61 LYGVGVEKTQLIMKSFTSPSNKKLEIQTVSSNYHIEMTPSDVGIYDRVVVQDLVKEMAQT 120
Query 133 RRDLPLGRH--KIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSR 190
+ + K+++L E DS+T AQ LRR ME +++ + L+C S S +IEPLQSR
Sbjct 121 SQIESTSQRSFKVVVLCEADSLTRDAQHGLRRTMEKYANNCKIVLSCESLSRIIEPLQSR 180
Query 191 CAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSAL 239
C I+ +D + + LR+V +E+ ++ ++ IV ++G++R A+
Sbjct 181 CIIINVPAPTDEDVTKVLRKVIERESFLLPENVLQKIVEKSEGNLRRAI 229
> Hs4506489
Length=356
Score = 106 bits (264), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 72/234 (30%), Positives = 112/234 (47%), Gaps = 44/234 (18%)
Query 42 SIWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARAL-- 99
S+W++KYRP +L + + E +LR + G+ PHLL+ GP G GK + ++C+ R L
Sbjct 2 SLWVDKYRPCSLGRLDYHKEQAAQLRNLVQCGDFPHLLVYGPSGAGKKTRIMCILRELYG 61
Query 100 -------------------------LQSRWRSCCLELNASDERSID--VVRDRIKAFAKE 132
+ S + LE+N SD + D V+++ +K A+
Sbjct 62 VGVEKLRIEHQTITTPSKKKIEISTIASNYH---LEVNPSDAGNSDRVVIQEMLKTVAQS 118
Query 133 R-------RDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIE 185
+ RD K+++L EVD +T+ AQ ALRR ME + T R L CNS+S VI
Sbjct 119 QQLETNSQRDF-----KVVLLTEVDKLTKDAQHALRRTMEKYMSTCRLILCCNSTSKVIP 173
Query 186 PLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSAL 239
P++SRC +R S + L VC KE + + + ++R AL
Sbjct 174 PIRSRCLAVRVPAPSIEDICHVLSTVCKKEGLNLPSQLAHRLAEKSCRNLRKAL 227
> ECU10g0780i
Length=354
Score = 103 bits (258), Expect = 4e-22, Method: Compositional matrix adjust.
Identities = 72/234 (30%), Positives = 113/234 (48%), Gaps = 37/234 (15%)
Query 43 IWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQS 102
IWIEKYRP++ DE++G +E L+ LE +PH+++ G G GK + +LCL L S
Sbjct 2 IWIEKYRPRSFDEMIGREEASGLLKSYTLE-TIPHMIVHGRSGHGKKTVLLCLVNHLYGS 60
Query 103 ----RWRSC------------------CLELNASDERSID--VVRDRIKAFAKER----- 133
+ R+ +E++ S D V++ IK + +
Sbjct 61 IPEMKIRTTEVLSGAKKIEVSYMESDEYVEISPSRYGYHDRAVIQSIIKEMGQTKPILSM 120
Query 134 ----RDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQS 189
R P+ K++++ + +T AQ ALRR +E++S R L CN S +IEP++S
Sbjct 121 LARSRRAPI---KLVVITSAEELTLEAQAALRRTIEVYSGVLRVVLVCNELSRLIEPIRS 177
Query 190 RCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQ 243
RC LR SD + + + KEN T + + I + GDMR AL L+
Sbjct 178 RCFFLRIPGFSDEDVTSNMCRILEKENYTVPKETLVEICRESGGDMRRALCVLE 231
> YBR087w
Length=354
Score = 102 bits (255), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 71/238 (29%), Positives = 127/238 (53%), Gaps = 35/238 (14%)
Query 42 SIWIEKYRPQALDEVVGNDEVLQRLRIIALEG-NMPHLLLAGPPGTGKTSSVLCL----- 95
S+W++KYRP++L+ + N+E+ L+ ++ + ++PHLLL GP GTGK + + L
Sbjct 2 SLWVDKYRPKSLNALSHNEELTNFLKSLSDQPRDLPHLLLYGPNGTGKKTRCMALLESIF 61
Query 96 ----------ARALLQSRWRSC---------CLELNASDERSID--VVRDRIKAFAK--- 131
R + + R LE+ SD + D V+++ +K A+
Sbjct 62 GPGVYRLKIDVRQFVTASNRKLELNVVSSPYHLEITPSDMGNNDRIVIQELLKEVAQMEQ 121
Query 132 ----ERRDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPL 187
+ +D R+K +I++E +S+T+ AQ ALRR ME +S R + C+S S +I P+
Sbjct 122 VDFQDSKDGLAHRYKCVIINEANSLTKDAQAALRRTMEKYSKNIRLIMVCDSMSPIIAPI 181
Query 188 QSRCAILRFTKLSDAHLVQRLREVCAKENVTF-TDDGIEAIVFSADGDMRSALNNLQS 244
+SRC ++R SD+ + L +V E + T D ++ I +++G++R +L L+S
Sbjct 182 KSRCLLIRCPAPSDSEISTILSDVVTNERIQLETKDILKRIAQASNGNLRVSLLMLES 239
> ECU05g1530
Length=568
Score = 92.8 bits (229), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 69/242 (28%), Positives = 121/242 (50%), Gaps = 15/242 (6%)
Query 8 NSSSAMNGSSSSSSSSGSSSSSSGSSSSSSSGNESIWIEKYRPQALDEVVGNDEVLQRLR 67
++S A++G++ + ++S S+SS +W EKYRP DE+VGN ++++L
Sbjct 81 DTSKALDGTAEVEKKTEKAASRDECGSASSG----VWSEKYRPSKRDEIVGNQGIVKQLE 136
Query 68 IIALEGNMPH--LLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDR 125
L+G + +LL+G PG GKT++ + ++L + +E NASD RS + ++
Sbjct 137 DY-LQGRTKYKAVLLSGQPGIGKTTTAHVVCKSLGLN-----VIEFNASDVRSKLEISNK 190
Query 126 IKAFAKERRDLPLG--RHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNS-SSS 182
+KAF + L G + K++I+DEVD M+ I + CN ++
Sbjct 191 VKAFVSSQSILRPGSSKSKVLIMDEVDGMSSDRGGIPELISIVKETVVPIICICNDRNNP 250
Query 183 VIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNL 242
I L S C LRF K ++ R++++ E D + I+ GD+R ++ +
Sbjct 251 KIRTLSSYCLDLRFRKPDARQILSRVKQILDMEGKKIPDGLLNEIISRGAGDIRYTISMV 310
Query 243 QS 244
QS
Sbjct 311 QS 312
> SPBC83.14c
Length=358
Score = 90.9 bits (224), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 87/351 (24%), Positives = 158/351 (45%), Gaps = 45/351 (12%)
Query 43 IWIEKYRPQALDEVVGNDEVLQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLARAL--- 99
+W+++YRP+ L + + ++ +RL ++ PHLL+ GP G GK + V+ + R L
Sbjct 2 LWLDQYRPKTLASLDYHKQLSERLISLSSTNEFPHLLVYGPSGAGKKTRVVAILRELYGP 61
Query 100 ----LQSRWRSCC-----------------LELNASDERSID--VVRDRIKAFAKERR-D 135
L+ R+ LE+ SD + D ++++ +K A+ + D
Sbjct 62 GSEKLKIDQRTFLTPSSKKLQINIVSSLHHLEITPSDVGNYDRVIMQELLKDVAQSAQVD 121
Query 136 LPLGR-HKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAIL 194
L + K+++++ D +T AQ ALRR ME +S+ R L NS+S +IEP++SR ++
Sbjct 122 LQAKKIFKVVVINVADELTRDAQAALRRTMEKYSNNIRLILIANSTSKIIEPIRSRTLMV 181
Query 195 RFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNK 254
R + ++ + ++ + + D + I + D ++R A+ L+ TV NK
Sbjct 182 RVAAPTPEEIILVMSKILTAQGLEAPDSLLNNIANNCDRNLRKAILLLE-TVHAKSPGNK 240
Query 255 ENVEKVCDTPPPEMLRRIMQQCVAGDWYGAQSVARV---------ILSLGYTPLDVVTTF 305
+ ++ P P+ I Q V QS AR+ +LS P ++
Sbjct 241 QLIDTGAQLPLPDWQTFIQQ--VGDSMLQEQSPARILAVRSMLYDLLSHCIPPTTILKEL 298
Query 306 RG-VLRRADSELQEHLLLEFLGIVGMTHMTMAGGLSTELQMEKMLAQLCKV 355
L + D++L +L+ H T G S +E +A KV
Sbjct 299 LSFFLSKVDTKLHPYLI---QAAANYEHRTRMGNKSI-FHLEAFVAYFMKV 345
> SPAC27E2.10c
Length=224
Score = 86.3 bits (212), Expect = 9e-17, Method: Compositional matrix adjust.
Identities = 56/212 (26%), Positives = 103/212 (48%), Gaps = 3/212 (1%)
Query 142 KIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSD 201
K+IILDE D+MT AAQ ALRR++E ++ RF + CN + + +QSRC RF L
Sbjct 4 KMIILDEADAMTLAAQNALRRVIEKYTKNVRFCIICNYINKISPAIQSRCTRFRFQPLPP 63
Query 202 AHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVC 261
+ + + V E+ D A++ + GDMR ALN LQ+ + + ++ +
Sbjct 64 KEIEKTVDHVIQSEHCNIDPDAKMAVLRLSKGDMRKALNILQACHAAYDHIDVSAIYNCV 123
Query 262 DTPPPEMLRRIMQQCVAGDWYGA-QSVARVILSLGYTPLDVVTTFRGVLRRADSELQEHL 320
P P + ++ + ++ A +++ + G D++T L + E++ +
Sbjct 124 GHPHPSDIDYFLKSIMNDEFVIAFNTISSIKQQKGLALQDILTCIFEALD--ELEIKPNA 181
Query 321 LLEFLGIVGMTHMTMAGGLSTELQMEKMLAQL 352
+ L + M+ G S ++Q+ M+A +
Sbjct 182 KIFILDQLATIEHRMSFGCSEKIQLSAMIASI 213
> At1g14460
Length=1102
Score = 79.7 bits (195), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 61/221 (27%), Positives = 101/221 (45%), Gaps = 36/221 (16%)
Query 46 EKYRPQALDEVVGNDEVLQRLRIIALEGNMPHL-LLAGPPGTGKTSSVLCLARAL----L 100
+KY+P DE++G V+Q L +G + H+ L GP GTGKTS+ L+ AL +
Sbjct 426 QKYKPMFFDELIGQSIVVQSLMNAVKKGRVAHVYLFQGPRGTGKTSTARILSAALNCDVV 485
Query 101 QSRWRSC----------------CLELNASDERSIDVVRDRIKAFAKERRDLPLG----- 139
+ C LEL+A + + VR +K L L
Sbjct 486 TEEMKPCGYCKECSDYMLGKSRDLLELDAGKKNGAEKVRYLLKKL------LTLAPQSSQ 539
Query 140 RHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSS--SVIEPLQSRCAILRFT 197
R+K+ ++DE + +L + +E + +F C ++ +V +QSRC F
Sbjct 540 RYKVFVIDECHLLPSRTWLSLLKFLE--NPLQKFVFVCITTDLDNVPRTIQSRCQKYIFN 597
Query 198 KLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSA 238
K+ D +V RLR++ + EN+ ++ I +ADG +R A
Sbjct 598 KVRDGDIVVRLRKIASDENLDVESQALDLIALNADGSLRDA 638
> At1g24290
Length=525
Score = 76.6 bits (187), Expect = 7e-14, Method: Compositional matrix adjust.
Identities = 75/254 (29%), Positives = 112/254 (44%), Gaps = 23/254 (9%)
Query 4 PTASNSSSAMNGSSSSSSSSGSSSSSSGSSSSSSSGNESIWIEKYRPQALDEVVGNDE-- 61
P + SS + + S+ SSSS E+ RP+ LD+VVG D
Sbjct 63 PAMGDDSSKRPNPKTQIHVAADDSNKRHKLSSSSHRQHQPLSERMRPRTLDDVVGQDHLL 122
Query 62 VLQRLRIIALEGN-MPHLLLAGPPGTGKTSSVLCLARALLQSR-----WRSCCLELNASD 115
L A+E N +P ++ GPPGTGKTS +A++L+ S +R L S
Sbjct 123 SPSSLLRSAVESNRLPSIVFWGPPGTGKTS----IAKSLINSSKDPSLYRFVSLSAVTS- 177
Query 116 ERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFAL 175
+ VRD ++ + +R +L + ++ +DEV ++ Q ++E S A
Sbjct 178 --GVKDVRDAVE--SAKRLNLEGRKRTVLFMDEVHRFNKSQQDTFLPVIEDGSILFIGAT 233
Query 176 ACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLR------EVCAKENVTFTDDGIEAIVF 229
N S +I PL SRC +L L H+ LR E +V D IE +
Sbjct 234 TENPSFHLITPLLSRCRVLTLNPLKPNHVETLLRRAVDDSERGLPNSVEVDDSVIEFLAN 293
Query 230 SADGDMRSALNNLQ 243
+ DGD R ALN L+
Sbjct 294 NCDGDARVALNALE 307
> 7297827
Length=318
Score = 74.3 bits (181), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 57/194 (29%), Positives = 94/194 (48%), Gaps = 28/194 (14%)
Query 79 LLAGPPGTGKTSSVLCLARALLQS---RWRS---------------------CCLELNAS 114
+ GP G GK + ++CL R + S R RS LE+N S
Sbjct 1 MFYGPSGAGKKTRIMCLLREMYGSGVERLRSETMTFTTPSNRKVEVMTVSSNYHLEVNPS 60
Query 115 DERSID--VVRDRIKAFAKERRDLPLGRH--KIIILDEVDSMTEAAQQALRRIMELHSDT 170
D D VV D IK A+ + G+ K+I++ E D +T+ AQ ALRR ME + T
Sbjct 61 DAGMYDRTVVIDLIKQVAQTHQIEISGQREFKVIVISEGDELTKDAQHALRRTMEKYVAT 120
Query 171 TRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFS 230
R ++ NS+S +I ++SRC +R ++ +V L+ C +E + + + +V
Sbjct 121 CRIIISVNSTSRIIPAIRSRCLGIRVAAPNETEIVSILQNTCKREGLALPVELAKRVVDK 180
Query 231 ADGDMRSALNNLQS 244
++ ++R AL L++
Sbjct 181 SERNLRRALLMLEA 194
> YMR078c
Length=741
Score = 73.9 bits (180), Expect = 5e-13, Method: Compositional matrix adjust.
Identities = 72/291 (24%), Positives = 122/291 (41%), Gaps = 75/291 (25%)
Query 20 SSSSGSSSSSSGSSSSSSSGNESIWIEKYRPQALDEVVGNDEV-------LQRLRIIALE 72
+S +++ S + G++++W+EK+RP+ ++VGN++ L++ +
Sbjct 92 ASGDDRTNAQKTSPITGKIGSDTLWVEKWRPKKFLDLVGNEKTNRRMLGWLRQWTPAVFK 151
Query 73 GNMPHL----------------------LLAGPPGTGKTSSVLCLARALLQSRWRSCCLE 110
+P L LL GPPG GKTS +A+ QS + E
Sbjct 152 EQLPKLPTEKEVSDMELDPLKRPPKKILLLHGPPGIGKTSVAHVIAK---QSGF--SVSE 206
Query 111 LNASDERSIDVVRDRIKAFAKERRDLPLGRHKI------IILDEVDSMTEAAQ-QALRRI 163
+NASDER+ +V+++I L H ++ DE+D E+ + L I
Sbjct 207 INASDERAGPMVKEKIYNL--------LFNHTFDTNPVCLVADEIDGSIESGFIRILVDI 258
Query 164 MELHSDTTRFALA-------------------------CNS-SSSVIEPLQSRCAILRFT 197
M+ T L CN+ + +E L+ C I+
Sbjct 259 MQSDIKATNKLLYGQPDKKDKKRKKKRSKLLTRPIICICNNLYAPSLEKLKPFCEIIAVK 318
Query 198 KLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSG 248
+ SD L++RL +C KEN+ I ++ A GD+R+ +NNLQ S
Sbjct 319 RPSDTTLLERLNLICHKENMNIPIKAINDLIDLAQGDVRNCINNLQFLASN 369
> At2g02480
Length=1153
Score = 69.7 bits (169), Expect = 9e-12, Method: Compositional matrix adjust.
Identities = 57/219 (26%), Positives = 94/219 (42%), Gaps = 33/219 (15%)
Query 46 EKYRPQALDEVVGNDEVLQRLRIIALEGNM-PHLLLAGPPGTGKTSSVLCLARAL---LQ 101
+KYRP +E++G V+Q L + P L GP GTGKTS+ + AL
Sbjct 453 QKYRPMFFEELIGQSIVVQSLMNAVKRSRIAPVYLFQGPRGTGKTSTARIFSAALNCVAT 512
Query 102 SRWRSCCL----------------ELNASDERSIDVVRDRIKAFAKERRDLP--LGR--- 140
+ C EL+ ++++ D VR +K +LP L R
Sbjct 513 EEMKPCGYCKECNDFMSGKSKDFWELDGANKKGADKVRYLLK-------NLPTILPRNSS 565
Query 141 -HKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKL 199
+K+ ++DE + + + +E F +V +QSRC F KL
Sbjct 566 MYKVFVIDECHLLPSKTWLSFLKFLENPLQKVVFIFITTDLENVPRTIQSRCQKFLFDKL 625
Query 200 SDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSA 238
D+ +V RL+++ + EN+ ++ I +ADG +R A
Sbjct 626 KDSDIVVRLKKIASDENLDVDLHALDLIAMNADGSLRDA 664
> At5g45720
Length=900
Score = 69.3 bits (168), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 67/280 (23%), Positives = 122/280 (43%), Gaps = 14/280 (5%)
Query 44 WIEKYRPQALDEVVGNDEVLQRL-RIIALEGNMPHLLLAGPPGTGKTSSVLCLARAL--- 99
+ +KY P+ +++G + V+Q L IA + GP GTGKTS ARAL
Sbjct 345 FTQKYAPRTFRDLLGQNLVVQALSNAIAKRRVGLLYVFHGPNGTGKTSCARVFARALNCH 404
Query 100 --LQSRWRSCCLELNASDERSIDVVRDR--IKAFAKERR-DLPLGRHK-----IIILDEV 149
QS+ C + D+ +R+ +K+F E D R + ++I D+
Sbjct 405 STEQSKPCGVCSSCVSYDDGKNRYIREMGPVKSFDFENLLDKTNIRQQQKQQLVLIFDDC 464
Query 150 DSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLR 209
D+M+ L +I++ F L C+S + + SRC F KL D ++ L+
Sbjct 465 DTMSTDCWNTLSKIVDRAPRRVVFVLVCSSLDVLPHIIVSRCQKFFFPKLKDVDIIDSLQ 524
Query 210 EVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGFGLVNKENVEKVCDTPPPEML 269
+ +KE + D ++ + +DG +R A L+ ++ V+++ E L
Sbjct 525 LIASKEEIDIDKDALKLVASRSDGSLRDAEMTLEQLSLLGTRISVPLVQEMVGLISDEKL 584
Query 270 RRIMQQCVAGDWYGAQSVARVILSLGYTPLDVVTTFRGVL 309
++ ++ D R+I+ G PL +++ V+
Sbjct 585 VDLLDLALSADTVNTVKNLRIIMETGLEPLALMSQLATVI 624
> At4g24790
Length=815
Score = 67.0 bits (162), Expect = 5e-11, Method: Compositional matrix adjust.
Identities = 56/211 (26%), Positives = 95/211 (45%), Gaps = 18/211 (8%)
Query 46 EKYRPQALDEVVGNDEVLQRLRIIALEGNMPHL-LLAGPPGTGKTSSVLCLARAL----- 99
+K+RP++ DE+VG + V++ L L G + + L GP GTGKTS+ A AL
Sbjct 243 QKFRPKSFDELVGQEVVVKCLLSTILRGRITSVYLFHGPRGTGKTSTSKIFAAALNCLSQ 302
Query 100 -LQSRWRSCCLELNAS-DERSIDVVR---------DRIKAFAKERRDLPLG-RHKIIILD 147
SR C E + R DV+ +++ K P+ R K+ I+D
Sbjct 303 AAHSRPCGLCSECKSYFSGRGRDVMETDSGKLNRPSYLRSLIKSASLPPVSSRFKVFIID 362
Query 148 EVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQR 207
E + + L ++ S + F L + + + SR F+K+ DA + +
Sbjct 363 ECQLLCQETWGTLLNSLDNFSQHSVFILVTSELEKLPRNVLSRSQKYHFSKVCDADISTK 422
Query 208 LREVCAKENVTFTDDGIEAIVFSADGDMRSA 238
L ++C +E + F ++ I +DG +R A
Sbjct 423 LAKICIEEGIDFDQGAVDFIASKSDGSLRDA 453
> YOR217w
Length=861
Score = 67.0 bits (162), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 59/251 (23%), Positives = 110/251 (43%), Gaps = 37/251 (14%)
Query 41 ESIWIEKYRPQALDEVVGNDEVLQRLRII------ALEGNMPHL-----------LLAGP 83
+ +W KY P L +V GN + +L+ + + + H +L GP
Sbjct 295 DKLWTVKYAPTNLQQVCGNKGSVMKLKNWLANWENSKKNSFKHAGKDGSGVFRAAMLYGP 354
Query 84 PGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIK------------AFAK 131
PG GKT++ +A+ L LE NASD RS ++ +K +
Sbjct 355 PGIGKTTAAHLVAQELGYD-----ILEQNASDVRSKTLLNAGVKNALDNMSVVGYFKHNE 409
Query 132 ERRDLPLGRHKIIILDEVDSMTEAAQQALRRIMEL-HSDTTRFALACNSSS-SVIEPLQS 189
E ++L G+H +II+DEVD M+ + + ++ + +T L CN + + P
Sbjct 410 EAQNLN-GKHFVIIMDEVDGMSGGDRGGVGQLAQFCRKTSTPLILICNERNLPKMRPFDR 468
Query 190 RCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSGF 249
C ++F + + RL + +E + I+ ++ + GD+R +N L + +
Sbjct 469 VCLDIQFRRPDANSIKSRLMTIAIREKFKLDPNVIDRLIQTTRGDIRQVINLLSTISTTT 528
Query 250 GLVNKENVEKV 260
+N EN+ ++
Sbjct 529 KTINHENINEI 539
> SPBC902.02c
Length=960
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 60/255 (23%), Positives = 106/255 (41%), Gaps = 58/255 (22%)
Query 41 ESIWIEKYRPQALDEVVGNDEV--------------------LQ---------RLRIIAL 71
+ +W++ YRPQ +++G++ V LQ R I
Sbjct 350 QKLWVDTYRPQLFRDLLGDERVHRAAMHWIKAWDPCVFGKSRLQPSKSMRFNPRFTNITS 409
Query 72 EGNMPH---LLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKA 128
+ + P ++L G G GKT+ +A Q+ ++ LE+NASD+R+ V +++ +
Sbjct 410 DSDRPDKRIMMLTGLAGAGKTTLAHVIAH---QAGYK--VLEINASDDRTAHTVHEKVSS 464
Query 129 FAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRFALACNSS-------- 180
L + +I+DE+D A +AL ++E T ++ A NS
Sbjct 465 AISNHSALS-SQPTCVIVDEIDGGDPAFVRALLSLLESDEKATEYSQAGNSKKKKKFKKL 523
Query 181 ------------SSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIV 228
+ + PL+ I+ F A LV RLR +C EN+ + +
Sbjct 524 CRPIICICNDLYTPALRPLRPYAQIIYFRPPPQASLVGRLRTICRNENIAVDSRSLTLLT 583
Query 229 FSADGDMRSALNNLQ 243
+ D+RS +N+LQ
Sbjct 584 DIYNSDIRSCINSLQ 598
> 7296805
Length=986
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 53/236 (22%), Positives = 104/236 (44%), Gaps = 42/236 (17%)
Query 44 WIEKYRPQALDEVVGNDEVLQRLR---------IIALEGNM----PH------------- 77
W++K++P ++ E+VG + + +GN P+
Sbjct 423 WVDKHKPTSIKEIVGQAGAASNVTKLMNWLSKWYVNHDGNKKPQRPNPWAKNDDGSFYKA 482
Query 78 LLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFAKER---- 133
LL+GPPG GKT++ + + L +E NASD RS +++D + +
Sbjct 483 ALLSGPPGIGKTTTATLVVKELGFD-----AVEFNASDTRSKRLLKDEVSTLLSNKSLSG 537
Query 134 ----RDLPLGRHKIIILDEVDSMT-EAAQQALRRIMELHSDTT-RFALACNS-SSSVIEP 186
+ + R ++I+DEVD M + ++ ++ L D++ CN + I
Sbjct 538 YFTGQGQAVSRKHVLIMDEVDGMAGNEDRGGMQELIALIKDSSIPIICMCNDRNHPKIRS 597
Query 187 LQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNL 242
L + C LRF + + ++ +C KE V + +E I+ + + D+R ++N++
Sbjct 598 LVNYCYDLRFQRPRLEQIKGKIMSICFKEKVKISPAKVEEIIAATNNDIRQSINHI 653
> SPBC23E6.07c
Length=934
Score = 62.8 bits (151), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 55/238 (23%), Positives = 103/238 (43%), Gaps = 35/238 (14%)
Query 33 SSSSSSGNESIWIEKYRPQALDEVVGNDEVLQRLRIIALE-----------------GNM 75
S+ S IW KY P +L ++ GN V+Q+L+ + G
Sbjct 350 SNKKESQPSQIWTSKYAPTSLKDICGNKGVVQKLQKWLQDYHKNRKSNFNKPGPDGLGLY 409
Query 76 PHLLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKA------- 128
+LL+GPPG GKT++ +A+ L+ LELNASD RS ++ +++
Sbjct 410 KAVLLSGPPGIGKTTAAHLVAK--LEGY---DVLELNASDTRSKRLLDEQLFGVTDSQSL 464
Query 129 ---FAKERRDLPLGRHK-IIILDEVDSMTEAAQQALRRI-MELHSDTTRFALACNS-SSS 182
F + + + + + ++I+DE+D M+ + + ++ M + CN +
Sbjct 465 AGYFGTKANPVDMAKSRLVLIMDEIDGMSSGDRGGVGQLNMIIKKSMIPIICICNDRAHP 524
Query 183 VIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALN 240
+ PL LRF + + R+ + +E + + ++ +V DMR +N
Sbjct 525 KLRPLDRTTFDLRFRRPDANSMRSRIMSIAYREGLKLSPQAVDQLVQGTQSDMRQIIN 582
> SPAC26H5.02c
Length=497
Score = 61.2 bits (147), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 69/215 (32%), Positives = 99/215 (46%), Gaps = 26/215 (12%)
Query 46 EKYRPQALDEVVGNDEVLQR---LRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQS 102
E+ RP++LDE VG +E++ +R + + ++L G GTGKT+ LAR L+
Sbjct 88 ERARPKSLDEYVGQEELVGERGIIRNLIEQDRCNSMILWGSAGTGKTT----LAR-LIAV 142
Query 103 RWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPL-GRHKIIILDEVDSMTEAAQQALR 161
+S +E++A+ V D K F + L L GR II LDEV A Q
Sbjct 143 TTKSRFIEISATST----TVADCRKIFEDSQNYLTLTGRKTIIFLDEVHRFNRAQQDIFL 198
Query 162 RIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKE------ 215
++E T A N S + L SRC + KL+ ++ + L C E
Sbjct 199 PMVEKGLVTLIGATTENPSFRLNSALISRCPVFVLKKLTRDNVKKILNHACLLESERLGS 258
Query 216 ---NV-TFTDDGIEAIVFSADGDMRSALNNLQSTV 246
NV T D I AI DGD R ALN L+ ++
Sbjct 259 SMPNVETSIIDYISAI---TDGDARMALNALEMSI 290
> At4g18820
Length=1057
Score = 60.5 bits (145), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 54/218 (24%), Positives = 92/218 (42%), Gaps = 33/218 (15%)
Query 46 EKYRPQALDEVVGNDEVLQRLRIIALEGNMPHL-LLAGPPGTGKTSSVLCLARAL----- 99
EKY P+ +++G + V+Q L + L + GP GTGKTS ARAL
Sbjct 435 EKYTPKTFRDLLGQNLVVQALSNAVARRKLGLLYVFHGPNGTGKTSCARIFARALNCHSM 494
Query 100 ----------------LQSRW--RSCCLELNASDERSIDVVRDRIKAFAKERRDLPLGRH 141
+ W R N E+ +D++ + ++ R
Sbjct 495 EQPKPCGTCSSCVSHDMGKSWNIREVGPVGNYDFEKIMDLLDGNVMVSSQSPR------- 547
Query 142 KIIILDEVDSMTEAAQQALRRIMELHSD-TTRFALACNSSSSVIEPLQSRCAILRFTKLS 200
+ I D+ D+++ AL ++++ + F L C+S + + SRC F KL
Sbjct 548 -VFIFDDCDTLSSDCWNALSKVVDRAAPRHVVFILVCSSLDVLPHVIISRCQKFFFPKLK 606
Query 201 DAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSA 238
DA +V L+ + +KE + D ++ I +DG +R A
Sbjct 607 DADIVYSLQWIASKEEIEIDKDALKLIASRSDGSLRDA 644
> Hs22045214
Length=144
Score = 57.4 bits (137), Expect = 4e-08, Method: Compositional matrix adjust.
Identities = 34/114 (29%), Positives = 50/114 (43%), Gaps = 23/114 (20%)
Query 189 SRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRSALNNLQSTVSG 248
S+C +LR+TKL+DA R K V +TD G+EAI+ A R
Sbjct 40 SQCIVLRYTKLTDARTSVRPMNAREKGTVLYTDFGLEAIILMARSMFR------------ 87
Query 249 FGLVNKENVEKVCDTPPPEMLRRIMQQCVAGDWYGAQSVARVILSLGYTPLDVV 302
VCD P P +++ ++Q CV + + LGY+P DV+
Sbjct 88 -----------VCDEPHPLLMKEMIQHCVNAHVKETYKIPAHLWHLGYSPEDVI 130
> Hs14777079
Length=1003
Score = 57.0 bits (136), Expect = 6e-08, Method: Compositional matrix adjust.
Identities = 52/192 (27%), Positives = 81/192 (42%), Gaps = 32/192 (16%)
Query 79 LLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNASDERSIDVVRDRIKAFAKERRDLPL 138
LL GPPG GKT+ +AR S +E+NASD+RS +V R RI+A + L
Sbjct 399 LLCGPPGLGKTTLAHVIARHAGYS-----VVEMNASDDRSPEVFRTRIEAATQMESVLGA 453
Query 139 -GRHKIIILDEVDSMTEAAQQALRRIMELHSDTT-------------------------R 172
G+ +++DE+D AA L I+
Sbjct 454 GGKPNCLVIDEIDGAPVAAINVLLSILNRKGPQEVGPQGPAVPSGGGRRRRAEGGLLMRP 513
Query 173 FALACNSS-SSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSA 231
CN + + L+ + +L F + LVQRL+EV ++ + + A+
Sbjct 514 IICICNDQFAPSLRQLKQQAFLLHFPPTLPSRLVQRLQEVSLRQGMRADPGVLAALCEKT 573
Query 232 DGDMRSALNNLQ 243
D D+R+ +N LQ
Sbjct 574 DNDIRACINTLQ 585
> CE20613
Length=839
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 60/253 (23%), Positives = 111/253 (43%), Gaps = 53/253 (20%)
Query 44 WIEKYRPQALDEVVGND-------EVLQRLRIIAL----EG------------------N 74
W++KY+P+ + E+VG + ++L+ ++ A EG +
Sbjct 280 WVDKYKPKRMGELVGQNGDKSPINKLLEWIKDWAKHNLGEGAKIKKPKPAPWMSSQDGTS 339
Query 75 MPHLLLAGPPGTGKTSSVLCLARAL-LQSRWRSCCLELNASDERSIDVVRDRI------- 126
LL+G PG GKT+ + L LQ +E+NASD RS + +I
Sbjct 340 FKAALLSGSPGVGKTTCAYMACQQLGLQ------LVEMNASDVRSKKHLEAKIGELSGSH 393
Query 127 --KAFAKERRDLPLGR---HKIIILDEVDSMTEAAQQA-LRRIMELHSDTTRFALAC--- 177
+ F ++ P H I+I+DEVD M+ +A + ++++ ++ + + C
Sbjct 394 QIEQFFGAKKCAPQDNQKVHHILIMDEVDGMSGNEDRAGISELIQIIKES-KIPIICICN 452
Query 178 NSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDDGIEAIVFSADGDMRS 237
+ I L + C LRF K + R+ +C++E + + ++ I+ + D+R
Sbjct 453 DRQHPKIRTLANYCYDLRFPKPRVETIRSRMMTICSQEKMKIGKEELDEIIELSGHDVRQ 512
Query 238 ALNNLQSTVSGFG 250
+ NLQ G G
Sbjct 513 TIYNLQMRSKGVG 525
> Hs18426902
Length=665
Score = 55.8 bits (133), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 60/217 (27%), Positives = 93/217 (42%), Gaps = 34/217 (15%)
Query 55 EVVGNDEVLQRLRIIALEGN-MPHLLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNA 113
+ VG D +L+ L LE N +P L+L GPPG GKT+ +A + R + L+A
Sbjct 244 KAVGQDTLLRSL----LETNEIPSLILWGPPGCGKTTLAHIIASNSKKHSIR--FVTLSA 297
Query 114 SDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRIMELHSDTTRF 173
++ ++ D VRD IK E+ R I+ +DE+ ++ Q +E + T
Sbjct 298 TNAKTND-VRDVIKQAQNEKSF--FKRKTILFIDEIHRFNKSQQDTFLPHVECGTITLIG 354
Query 174 ALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVC--------------------- 212
A N S V L SRC ++ KL +V L
Sbjct 355 ATTENPSFQVNAALLSRCRVIVLEKLPVEAMVTILMRAINSLGIHVLDSSRPTDPLSHSS 414
Query 213 --AKENVTFTDD-GIEAIVFSADGDMRSALNNLQSTV 246
+ E F +D ++ + + +DGD R+ LN LQ V
Sbjct 415 NSSSEPAMFIEDKAVDTLAYLSDGDARAGLNGLQLAV 451
> Hs15011931
Length=1148
Score = 55.5 bits (132), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 64/263 (24%), Positives = 107/263 (40%), Gaps = 49/263 (18%)
Query 22 SSGSSSSSSGSSSSSSSGNESI-WIEKYRPQALDEVVGN-------DEVLQRLR------ 67
+SG S + + + SS + E++ W++KY+P +L ++G +++L+ LR
Sbjct 563 TSGDSKARNLADDSSENKVENLLWVDKYKPTSLKTIIGQQGDQSCANKLLRWLRNWQKSS 622
Query 68 ---------IIALEGN-----MPHLLLAGPPGTGKTSSVLCLARALLQSRWRSCCLELNA 113
G LL+GPPG GKT++ + + L S +ELNA
Sbjct 623 SEDKKHAAKFGKFSGKDDGSSFKAALLSGPPGVGKTTTASLVCQELGYS-----YVELNA 677
Query 114 SDERSIDVVR---------DRIKAFAKERRDLPLGRHKIIILDEVDSMTEAAQQALRRIM 164
SD RS ++ IK F + +I+DEVD M A + I
Sbjct 678 SDTRSKSSLKAIVAESLNNTSIKGFYSNGAASSVSTKHALIMDEVDGM--AGNEDRGGIQ 735
Query 165 EL-----HSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTF 219
EL H+ + + + I L C LRF + + + + KE +
Sbjct 736 ELIGLIKHTKIPIICMCNDRNHPKIRSLVHYCFDLRFQRPRVEQIKGAMMSIAFKEGLKI 795
Query 220 TDDGIEAIVFSADGDMRSALNNL 242
+ I+ A+ D+R L+NL
Sbjct 796 PPPAMNEIILGANQDIRQVLHNL 818
> ECU01g1180
Length=383
Score = 52.4 bits (124), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 56/209 (26%), Positives = 93/209 (44%), Gaps = 33/209 (15%)
Query 42 SIWIEKYRPQALDEVVGNDEV----LQRLRIIALEGNMPHLLLAGPPGTGKTSSVLCLAR 97
++W EKYRP+ E+V +V L+ LR G + LLL GPPGTGKT+ LA
Sbjct 4 ALWTEKYRPRKYTELVFKGDVHHDALRWLRDYPSNGRI--LLLCGPPGTGKTA----LAH 57
Query 98 ALLQSRWRSCCLELNA-SDERSIDVVRDRIKAFAKERRDLPLGRHKIIILDEVDS--MTE 154
+L S + +E NA SD + + D G+ +I++DE+D + E
Sbjct 58 -VLSSVFGMNLVEFNASSDSEYVSKILDSGGTIN--------GKKNLILVDEIDGNPLIE 108
Query 155 AAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAK 214
+ A +TR A +S+ E L + + + + +C +
Sbjct 109 VGRLA---------SSTRLAHPVVMTSN--EVRLKDVYTLEIKRPGIDEIRRGIERICRE 157
Query 215 ENVTFTDDGIEAIVFSADGDMRSALNNLQ 243
E + + + +V + GD R+ +N+LQ
Sbjct 158 EGIRVDNSVLTRMVEDSGGDFRAIINHLQ 186
> Hs22058826
Length=118
Score = 49.3 bits (116), Expect = 1e-05, Method: Composition-based stats.
Identities = 25/74 (33%), Positives = 38/74 (51%), Gaps = 0/74 (0%)
Query 163 IMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLVQRLREVCAKENVTFTDD 222
++E ++ TRF L CN S +I LQSRC RF L+ +V RL V +E +
Sbjct 14 VIEKFTENTRFCLICNYLSKIIPALQSRCTRFRFGPLTPELMVPRLEHVVEEEKKNPGQE 73
Query 223 GIEAIVFSADGDMR 236
I + + DG+ +
Sbjct 74 RIVELFLAVDGESK 87
> YNL218w
Length=587
Score = 48.5 bits (114), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 62/230 (26%), Positives = 94/230 (40%), Gaps = 45/230 (19%)
Query 46 EKYRPQALDEVVGNDEVLQR----LRIIALEGNMPHLLLAGPPGTGKTSSVLCLARALLQ 101
EK RP+ L + VG +L + L +G +P ++L GPPG GKTS LAR L +
Sbjct 137 EKLRPKELRDYVGQQHILSQDNGTLFKYIKQGTIPSMILWGPPGVGKTS----LARLLTK 192
Query 102 ------------SRW---RSCCLELNASDERSIDVVRDRIKAFAKERRDLPL-GRHKIII 145
SR+ + + N + R I F K +++ L R ++
Sbjct 193 TATTSSNESNVGSRYFMIETSATKANTQELRGI---------FEKSKKEYQLTKRRTVLF 243
Query 146 LDEVDSMTEAAQQALRRIMELHSDTTRFALACNSSSSVIEPLQSRCAILRFTKLSDAHLV 205
+DE+ + Q L +E A N S + L SRC I KL+ L
Sbjct 244 IDEIHRFNKVQQDLLLPHVENGDIILIGATTENPSFQLNNALISRCLIFVLEKLNVNELC 303
Query 206 QRL-----------REVCAKEN-VTFTDDGIEAIVFSADGDMRSALNNLQ 243
L ++V EN + + +E +V + GD R ALN L+
Sbjct 304 IVLSRGIALLNKCRKQVWNIENPLKLSRSILEYVVDLSVGDTRRALNMLE 353
Lambda K H
0.316 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 8945217930
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40