bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
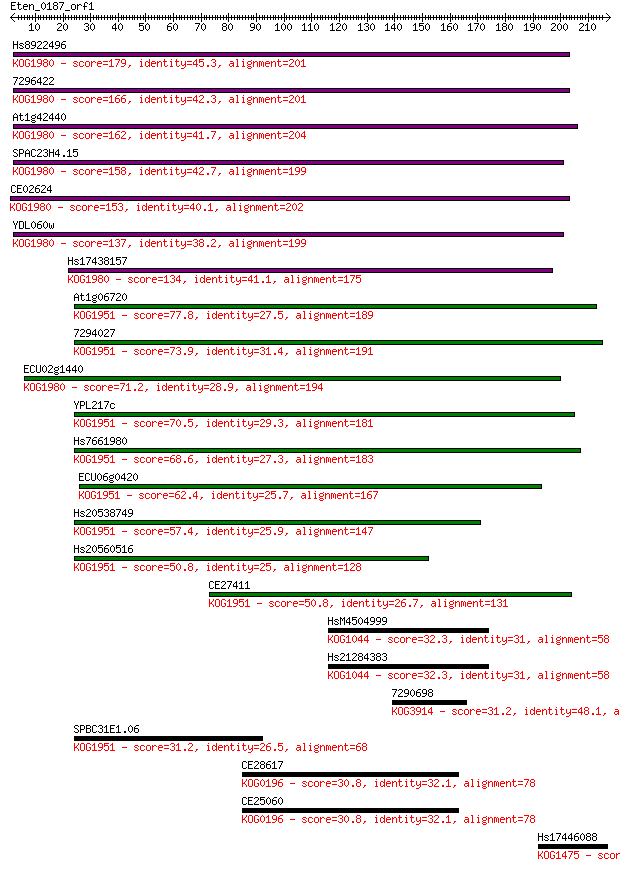
Query= Eten_0187_orf1
Length=217
Score E
Sequences producing significant alignments: (Bits) Value
Hs8922496 179 3e-45
7296422 166 3e-41
At1g42440 162 7e-40
SPAC23H4.15 158 6e-39
CE02624 153 2e-37
YDL060w 137 1e-32
Hs17438157 134 1e-31
At1g06720 77.8 1e-14
7294027 73.9 2e-13
ECU02g1440 71.2 1e-12
YPL217c 70.5 2e-12
Hs7661980 68.6 8e-12
ECU06g0420 62.4 8e-10
Hs20538749 57.4 2e-08
Hs20560516 50.8 2e-06
CE27411 50.8 2e-06
HsM4504999 32.3 0.83
Hs21284383 32.3 0.83
7290698 31.2 1.5
SPBC31E1.06 31.2 1.6
CE28617 30.8 2.2
CE25060 30.8 2.3
Hs17446088 30.8 2.3
> Hs8922496
Length=656
Score = 179 bits (454), Expect = 3e-45, Method: Compositional matrix adjust.
Identities = 91/203 (44%), Positives = 134/203 (66%), Gaps = 11/203 (5%)
Query 2 LLQHETQVAVVHSQLTF--CDDQPIKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKW 59
LL HE +++V++ + + +P+K+K+ ++ CGFRRF ASP++S+ +A K
Sbjct 436 LLPHEQKMSVLNMVVRRDPGNTEPVKAKEELIFHCGFRRFRASPLFSQHT-----AADKH 490
Query 60 KFKRWAEPGSTLTATVYSPLVLPPSPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLII 119
K +R+ L ATVY+P+ PP+ ++ + S+ +L A G ++ V+ R++I
Sbjct 491 KLQRFLTADMALVATVYAPITFPPASVLLFKQKSNGMHSL--IATGHLMSVD--PDRMVI 546
Query 120 KRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTHGYMKCK 179
KRV+LSGHPFK+ + A+VR+MFFN +DV WF P+EL TK G RG+I EPLGTHG+MKC
Sbjct 547 KRVVLSGHPFKIFTKMAVVRYMFFNREDVLWFKPVELRTKWGRRGHIKEPLGTHGHMKCS 606
Query 180 FSDHLKQSDEVLLPLYRRVFPKW 202
F LK D VL+ LY+RVFPKW
Sbjct 607 FDGKLKSQDTVLMNLYKRVFPKW 629
> 7296422
Length=814
Score = 166 bits (420), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 85/204 (41%), Positives = 129/204 (63%), Gaps = 13/204 (6%)
Query 2 LLQHETQVAVVHSQLTFCDDQ--PIKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKW 59
+L HE Q+ V++ L D P+KSK+ +++ CG+RRF +PIYS+ + K
Sbjct 590 MLPHEHQMCVMNVVLQRMPDSEVPLKSKEQLIIQCGYRRFVVNPIYSQHT-----NGDKH 644
Query 60 KFKRWAEPGSTLTATVYSPLVLPPSPCIMLRSDSSEPDN-LRITAWGSVLPVNGASSRLI 118
KF+R+ P T+ AT Y+P+ PP+P + + + PD+ L + A G +L N R++
Sbjct 645 KFERYFRPYETVCATFYAPIQFPPAPVLAFKVN---PDSTLALVARGRLLSCN--PDRIV 699
Query 119 IKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTHGYMKC 178
+KRV+LSGHP +++R+ A +R+MFF +DV +F P++L TK G G+I E LGTHG+MKC
Sbjct 700 LKRVVLSGHPMRINRKSASIRYMFFYKEDVEYFKPVKLRTKCGRLGHIKESLGTHGHMKC 759
Query 179 KFSDHLKQSDEVLLPLYRRVFPKW 202
F L+ D + LY+RVFPKW
Sbjct 760 YFDGQLRSYDTAFMYLYKRVFPKW 783
> At1g42440
Length=766
Score = 162 bits (409), Expect = 7e-40, Method: Compositional matrix adjust.
Identities = 85/211 (40%), Positives = 138/211 (65%), Gaps = 15/211 (7%)
Query 2 LLQHETQVAVVHSQLTFCD--DQPIKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKW 59
LLQHE++++V+H + D + PIK+K+ ++ GFR+F A P+++ + S+ K
Sbjct 562 LLQHESKMSVLHFSVKKYDGYEAPIKTKEELMFHVGFRQFIARPVFATDNF----SSDKH 617
Query 60 KFKRWAEPGSTLTATVYSPLVLPPSPCIMLR-SDSSEPDNLRITAWGSVLPVNGASSRLI 118
K +R+ PG A++Y P+ PP P ++L+ S+ S+P I A GS+ V +++I
Sbjct 618 KMERFLHPGCFSLASIYGPISFPPLPLVVLKISEGSDPPA--IAALGSLKSVE--PNKII 673
Query 119 IKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTHGYMKC 178
+K+++L+G+P +V + KA VR+MF NP+DV+WF P+E+ +K G RG + EP+GTHG MKC
Sbjct 674 LKKIILTGYPQRVSKMKASVRYMFHNPEDVKWFKPVEVWSKCGRRGRVKEPVGTHGAMKC 733
Query 179 KFSDHLKQSDEVLLPLYRRVFPKW----YPQ 205
F+ ++Q D V + LY+R +PKW YPQ
Sbjct 734 IFNGVVQQHDVVCMNLYKRAYPKWPERLYPQ 764
> SPAC23H4.15
Length=783
Score = 158 bits (400), Expect = 6e-39, Method: Compositional matrix adjust.
Identities = 85/203 (41%), Positives = 125/203 (61%), Gaps = 11/203 (5%)
Query 2 LLQHETQVAVVHSQLTFCD----DQPIKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAA 57
LLQ+E ++ V SQ T ++PI+SK+ +L+ G RRF P+YS+ G +
Sbjct 575 LLQYENKLTV--SQFTAMQHSEYEEPIESKEELLLQIGPRRFMVRPLYSDPTASGASNNL 632
Query 58 KWKFKRWAEPGSTLTATVYSPLVLPPSPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRL 117
+ K+ R+ P + A+V SP+V P IM + S ++LR+ A GS VN ++ +
Sbjct 633 Q-KYHRYLPPKQAVIASVISPIVFGNVPIIMFKKSSD--NSLRLAATGSY--VNCDTNSV 687
Query 118 IIKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTHGYMK 177
I KR +L+GHPFKVH++ +R+MFFNP+DV WF PI+L TK+G G I EPLGTHGY K
Sbjct 688 IAKRAVLTGHPFKVHKKLVTIRYMFFNPEDVIWFKPIQLFTKQGRTGYIKEPLGTHGYFK 747
Query 178 CKFSDHLKQSDEVLLPLYRRVFP 200
F+ + D V + LY+R++P
Sbjct 748 ATFNGKITVQDTVAMSLYKRMYP 770
> CE02624
Length=785
Score = 153 bits (387), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 81/210 (38%), Positives = 118/210 (56%), Gaps = 21/210 (10%)
Query 1 QLLQHETQVAVVHSQLT--------FCDDQPIKSKDPMLMACGFRRFPASPIYSEEPRQG 52
QLL HE +++V++ L DQ K + GFR+F A+ ++S
Sbjct 556 QLLPHEHKMSVLNMVLKKHPSCTIPITSDQNQK----FIFYVGFRQFEANAVFSSNT--- 608
Query 53 VRSAAKWKFKRWAEPGSTLTATVYSPLVLPPSPCIMLRSDSSEPDNLRITAWGSVLPVNG 112
K+K +R+ T ATVY+P+ P+ + R D L A GS+L N
Sbjct 609 --PGDKFKLERFMPTEKTFVATVYAPITFNPATVLCFRQDDKGRQEL--VATGSILDSN- 663
Query 113 ASSRLIIKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGT 172
R+++KR +L+GHP+K++RR +VR+MFFN +D+ WF P+EL+T G RG+I E +GT
Sbjct 664 -PDRIVLKRTVLAGHPYKINRRAVVVRYMFFNREDIEWFKPVELYTPSGRRGHIKEAVGT 722
Query 173 HGYMKCKFSDHLKQSDEVLLPLYRRVFPKW 202
HG MKC+F L D V+L LY+RVFP W
Sbjct 723 HGNMKCRFDQQLNAQDSVMLNLYKRVFPVW 752
> YDL060w
Length=788
Score = 137 bits (346), Expect = 1e-32, Method: Compositional matrix adjust.
Identities = 76/202 (37%), Positives = 113/202 (55%), Gaps = 9/202 (4%)
Query 2 LLQHETQVAVVHSQLTFCD--DQPIKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAK- 58
LL HE + AVV+ L + D+P+ S++P+++ G RR+ P++S QG S
Sbjct 584 LLLHEHKNAVVNFSLQRWEQYDKPVPSQEPIVVQYGVRRYTIQPLFS----QGSNSPNNV 639
Query 59 WKFKRWAEPGSTLTATVYSPLVLPPSPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLI 118
K++R+ P + AT +P+ SP I + ++ N+ + G+ L N SR++
Sbjct 640 HKYERFLHPDTVSVATCIAPVDFTQSPAIFFKPSPTDAKNIELIGHGTFL--NADHSRIL 697
Query 119 IKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTHGYMKC 178
KR +L+GHPF+ H+ VR+MFF P+DV WF I L TK G G I E LGTHGY K
Sbjct 698 AKRAILTGHPFRFHKTVVTVRYMFFRPEDVEWFKSIPLFTKSGRSGFIKESLGTHGYFKA 757
Query 179 KFSDHLKQSDEVLLPLYRRVFP 200
F L D V + LY+R++P
Sbjct 758 TFDGKLSAQDVVAMSLYKRMWP 779
> Hs17438157
Length=465
Score = 134 bits (337), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 72/175 (41%), Positives = 105/175 (60%), Gaps = 9/175 (5%)
Query 22 QPIKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVL 81
+ K+K+ ++ C R F ASP++S+ +A K KF+R+ L TVY+P
Sbjct 300 ESAKAKEELIFRCRSRHFRASPLFSQHA-----AADKHKFQRFLTADMALAVTVYAPTTF 354
Query 82 PPSPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRFM 141
PPS ++ + S+ NL A G +L V+ +R++IKRV+LS PFK+ + A+V +M
Sbjct 355 PPSSLLLFKQKSNGMHNL--IATGHLLLVD--PNRMVIKRVVLSDPPFKICTKMAVVCYM 410
Query 142 FFNPDDVRWFTPIELHTKRGLRGNIVEPLGTHGYMKCKFSDHLKQSDEVLLPLYR 196
FFN +DV+WF P+EL TK RG+I EP GTHG MKC F LK + L+ LY+
Sbjct 411 FFNREDVQWFKPVELRTKWSRRGHIQEPSGTHGRMKCSFDRKLKSQNTELMNLYK 465
> At1g06720
Length=1147
Score = 77.8 bits (190), Expect = 1e-14, Method: Composition-based stats.
Identities = 52/207 (25%), Positives = 100/207 (48%), Gaps = 25/207 (12%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPP 83
+K++DP++++ G+RR+ P+++ E R G + + ++ A+ + PLV P
Sbjct 806 LKTRDPIIVSIGWRRYQTIPVFAIEDRNG-----RHRMLKYTPEHMHCLASFWGPLVPPN 860
Query 84 SPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRFMFF 143
+ + ++ S+ RITA VL N + I+K++ L G P K+ ++ A ++ MF
Sbjct 861 TGFVAFQNLSNNQAGFRITATSVVLEFNHQAR--IVKKIKLVGTPCKIKKKTAFIKDMFT 918
Query 144 NPDDVRWFTPIELHTKRGLRG-------NIVEPLGTHGYMKCKFSDHLKQSDEVLLPLYR 196
+ ++ F + T G+RG N+++ G +C F D + SD V L +
Sbjct 919 SDLEIARFEGSSVRTVSGIRGQVKKAGKNMLDNKAEEGIARCTFEDQIHMSDMVFLRAWT 978
Query 197 RV-FPKWY----------PQSWGGLPT 212
V P++Y ++W G+ T
Sbjct 979 TVEVPQFYNPLTTALQPRDKTWNGMKT 1005
> 7294027
Length=1159
Score = 73.9 bits (180), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 60/199 (30%), Positives = 100/199 (50%), Gaps = 15/199 (7%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPP 83
+K+ DP++++ G+RRF IY++ V + ++ ++ T + T + P+
Sbjct 821 LKTGDPLIISMGWRRFQTVAIYAK-----VEDNFRQRYLKYTPNHVTCSMTFWGPITPQN 875
Query 84 SPCIML---RSDSSEPDNL--RITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIV 138
+ + L R D E L RI A G V V+ +S I+K++ L GHPFK++++ A V
Sbjct 876 TGFLALQTVRQDQEEMRRLGFRIAATGCVTEVDKSSQ--IMKKLKLVGHPFKIYKKTAFV 933
Query 139 RFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGT-HGYMKCKFSDHLKQSDEVLLPLYRR 197
+ MF + +V F ++ T G+RG I + T G + F D + SD V + R
Sbjct 934 KDMFNSSLEVAKFEGAKIKTVSGIRGQIKKAHHTPEGSYRATFEDKILLSDIVFCRTWFR 993
Query 198 V-FPKWY-PQSWGGLPTQQ 214
V P++Y P S LP Q
Sbjct 994 VEVPRFYAPVSSLLLPLDQ 1012
> ECU02g1440
Length=544
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 56/205 (27%), Positives = 97/205 (47%), Gaps = 21/205 (10%)
Query 6 ETQVAVVHSQLTFCDDQPIKS----------KDPMLMACGFRRFPASPIYSEEPRQGVRS 55
E Q+ VV + D + I + ++ M++ G + P+ ++ Q V
Sbjct 346 EEQIYVVFGCYEYEDRKTIHNFHFEGEVPLKEESMVVDLGHKIVDICPMITKNLNQKV-- 403
Query 56 AAKWKFKRWAEPGSTLTATVYSPLVLPPSPCIMLRSDSSEPDNLRITAWGSVLPVNGASS 115
FKR E S + + + P+ S ++ R S L T SV
Sbjct 404 -----FKRQKELESGVISFI-GPISFGLSRVLIYRK--SALRELSATTLASVGMNGFMGD 455
Query 116 RLIIKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTK-RGLRGNIVEPLGTHG 174
R+I + +L G PFK +R ++V+ MF + ++V +F I+++ + R + G I +P+GT G
Sbjct 456 RVIFEEAVLQGIPFKNKKRYSLVKRMFNSKEEVMYFRNIQIYMRNRKITGFIKKPIGTKG 515
Query 175 YMKCKFSDHLKQSDEVLLPLYRRVF 199
K FS +K D+V++ LY+RVF
Sbjct 516 TFKGYFSQPIKSGDKVMMSLYKRVF 540
> YPL217c
Length=1183
Score = 70.5 bits (171), Expect = 2e-12, Method: Composition-based stats.
Identities = 53/185 (28%), Positives = 89/185 (48%), Gaps = 19/185 (10%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTL-TATVYSPLVLP 82
+K+ DP++++ G+RRF PIY+ + K+ P T A Y PL P
Sbjct 839 LKTNDPLVLSLGWRRFQTLPIYTTTDSRTRTRMLKYT------PEHTYCNAAFYGPLCSP 892
Query 83 PSP-C-IMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRF 140
+P C + + ++S + RI A G V ++ + I+K++ L G P+K+ + A ++
Sbjct 893 NTPFCGVQIVANSDTGNGFRIAATGIVEEID--VNIEIVKKLKLVGFPYKIFKNTAFIKD 950
Query 141 MFFNPDDVRWFTPIELHTKRGLRGNIVEPLGT-HGYMKCKFSDHLKQSDEVLLPLYRRVF 199
MF + +V F ++ T G+RG I L G+ + F D + SD V+L
Sbjct 951 MFSSAMEVARFEGAQIKTVSGIRGEIKRALSKPEGHYRAAFEDKILMSDIVIL------- 1003
Query 200 PKWYP 204
WYP
Sbjct 1004 RSWYP 1008
> Hs7661980
Length=1282
Score = 68.6 bits (166), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 50/184 (27%), Positives = 87/184 (47%), Gaps = 15/184 (8%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPP 83
+KS+DP++ + G+RRF P+Y E G + + ++ A + P+
Sbjct 936 LKSRDPIIFSVGWRRFQTIPLYYIEDHNG-----RQRLLKYTPQHMHCGAAFWGPITPQG 990
Query 84 SPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRFMFF 143
+ + ++S S + RI A G VL ++ S I+K++ L+G P+K+ + + ++ MF
Sbjct 991 TGFLAIQSVSGIMPDFRIAATGVVLDLD--KSIKIVKKLKLTGFPYKIFKNTSFIKGMFN 1048
Query 144 NPDDVRWFTPIELHTKRGLRGNIVEPL-GTHGYMKCKFSDHLKQSDEVLLPLYRRVFPKW 202
+ +V F + T G+RG I + L G + F D L SD V + W
Sbjct 1049 SALEVAKFEGAVIRTVSGIRGQIKKALRAPEGAFRASFEDKLLMSDIVFMRT-------W 1101
Query 203 YPQS 206
YP S
Sbjct 1102 YPVS 1105
> ECU06g0420
Length=777
Score = 62.4 bits (150), Expect = 8e-10, Method: Composition-based stats.
Identities = 43/167 (25%), Positives = 79/167 (47%), Gaps = 12/167 (7%)
Query 26 SKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPPSP 85
+ +P + + G+RRF + P++S ++ A + + ++ + Y P+V P
Sbjct 464 TNEPAIFSVGWRRFQSIPVFS------MKDATRNRMIKYTPESMHCNVSFYGPVV----P 513
Query 86 CIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRFMFFNP 145
S SE + R+ A G++ VNG + ++K++ L G+P ++ + VR MF +
Sbjct 514 AGTGFSVYSEKGDFRVLALGTITDVNGDAK--LVKKLKLVGYPKQIVQNTVFVRDMFTSD 571
Query 146 DDVRWFTPIELHTKRGLRGNIVEPLGTHGYMKCKFSDHLKQSDEVLL 192
+V F L GLRG + P G +G + F + SD + L
Sbjct 572 LEVLKFEGASLKAVSGLRGQVKGPHGKNGEYRAVFEGKMLMSDIITL 618
> Hs20538749
Length=192
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 38/147 (25%), Positives = 72/147 (48%), Gaps = 7/147 (4%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPP 83
+KS+DP++ + G+RRF +Y E G + + ++ A + P+
Sbjct 53 LKSQDPIIFSVGWRRFQTILLYYIEDHNG-----RQRLLKYTPQHIHCGAAFWDPITPQG 107
Query 84 SPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRFMFF 143
+ + ++S S + RI A G VL ++ S I+K++ L+G P+K+ + + ++ MF
Sbjct 108 TGFLAIQSVSGIMPDFRIAATGVVLDLD--KSIKIVKKLKLTGFPYKIFKNTSFIKGMFN 165
Query 144 NPDDVRWFTPIELHTKRGLRGNIVEPL 170
+ +V F + T G+RG I L
Sbjct 166 SALEVAKFEDAVIRTVSGIRGQIKRAL 192
> Hs20560516
Length=227
Score = 50.8 bits (120), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/128 (25%), Positives = 65/128 (50%), Gaps = 7/128 (5%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPP 83
+KS+DP++ + G+RRF P+Y E G + + ++ A + P+
Sbjct 102 LKSRDPIIFSVGWRRFQTIPLYYIEDHNG-----RQRLLKYTPQHMHCGAAFWDPVTPQG 156
Query 84 SPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVHRRKAIVRFMFF 143
+ + ++S S + RI A G VL ++ S I+K++ L+G P+K+ + + ++ +F
Sbjct 157 TGFLAIQSVSGIMPDFRIAATGVVLDLD--KSIKIVKKLKLTGFPYKIFKNTSFIKGIFN 214
Query 144 NPDDVRWF 151
+ +V F
Sbjct 215 SALEVAKF 222
> CE27411
Length=920
Score = 50.8 bits (120), Expect = 2e-06, Method: Composition-based stats.
Identities = 35/133 (26%), Positives = 67/133 (50%), Gaps = 4/133 (3%)
Query 73 ATVYSPLVLPPSPCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKVH 132
A+ + P+ + + ++S + + RI A G VL ++ ++ ++K++ L GHP K+
Sbjct 619 ASFFGPVCAQNTGLLAIQSIADKTPGYRIVATGGVLDLDKSTQ--VVKKLKLIGHPEKIF 676
Query 133 RRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPL-GTHGYMKCKFSDHLKQSDEVL 191
++ A V+ MF + +V F + T G+RG I + + G + F D + D V
Sbjct 677 KKTAFVKGMFNSALEVAKFEGATIRTVAGIRGQIKKAIKAPEGAFRATFEDKILMRDIVF 736
Query 192 LPLYRRV-FPKWY 203
L + V P++Y
Sbjct 737 LRSWVTVPIPRFY 749
> HsM4504999
Length=778
Score = 32.3 bits (72), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 25/58 (43%), Gaps = 9/58 (15%)
Query 116 RLIIKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTH 173
RLI++ +SG F HRR I ++ P D + RG N V+P H
Sbjct 36 RLIVEDRRVSGTSFTAHRRATITHLLYLCPKD---------YCPRGRVCNSVDPFVAH 84
> Hs21284383
Length=778
Score = 32.3 bits (72), Expect = 0.83, Method: Compositional matrix adjust.
Identities = 18/58 (31%), Positives = 25/58 (43%), Gaps = 9/58 (15%)
Query 116 RLIIKRVLLSGHPFKVHRRKAIVRFMFFNPDDVRWFTPIELHTKRGLRGNIVEPLGTH 173
RLI++ +SG F HRR I ++ P D + RG N V+P H
Sbjct 36 RLIVEDRRVSGTSFTAHRRATITHLLYLCPKD---------YCPRGRVCNSVDPFVAH 84
> 7290698
Length=424
Score = 31.2 bits (69), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 13/27 (48%), Positives = 18/27 (66%), Gaps = 0/27 (0%)
Query 139 RFMFFNPDDVRWFTPIELHTKRGLRGN 165
R +F NPDD++ F IEL GL+G+
Sbjct 19 RVLFVNPDDLQIFKEIELPPDLGLKGH 45
> SPBC31E1.06
Length=838
Score = 31.2 bits (69), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 18/68 (26%), Positives = 32/68 (47%), Gaps = 5/68 (7%)
Query 24 IKSKDPMLMACGFRRFPASPIYSEEPRQGVRSAAKWKFKRWAEPGSTLTATVYSPLVLPP 83
+K+ DP++ + G+RRF + P+YS S + + ++ T Y P V P
Sbjct 776 LKTNDPLIFSMGWRRFQSIPVYSISD-----SRTRNRMLKYTPEHMHCFGTFYGPFVAPN 830
Query 84 SPCIMLRS 91
S ++S
Sbjct 831 SGFCAVQS 838
> CE28617
Length=1122
Score = 30.8 bits (68), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 38/90 (42%), Gaps = 14/90 (15%)
Query 85 PCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKV-HRRKAIVRFMFF 143
P LR D+S+ D + I AW SV + + + ++ ++ F+ H F+
Sbjct 451 PVSHLRVDASQSDGITI-AW-SVSDSDVSDFEVEVRPAIVKKRTFETRHVNMTYTTFIGL 508
Query 144 NP-----------DDVRWFTPIELHTKRGL 162
NP DD+RW PI RGL
Sbjct 509 NPETVYQFRVRIRDDLRWSQPISYQLGRGL 538
> CE25060
Length=1117
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 25/90 (27%), Positives = 38/90 (42%), Gaps = 14/90 (15%)
Query 85 PCIMLRSDSSEPDNLRITAWGSVLPVNGASSRLIIKRVLLSGHPFKV-HRRKAIVRFMFF 143
P LR D+S+ D + I AW SV + + + ++ ++ F+ H F+
Sbjct 451 PVSHLRVDASQSDGITI-AW-SVSDSDVSDFEVEVRPAIVKKRTFETRHVNMTYTTFIGL 508
Query 144 NP-----------DDVRWFTPIELHTKRGL 162
NP DD+RW PI RGL
Sbjct 509 NPETVYQFRVRIRDDLRWSQPISYQLGRGL 538
> Hs17446088
Length=267
Score = 30.8 bits (68), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 13/25 (52%), Positives = 16/25 (64%), Gaps = 0/25 (0%)
Query 192 LPLYRRVFPKWYPQSWGGLPTQQPL 216
+P+ RR FPK Q+WG PTQ L
Sbjct 1 MPMTRRGFPKAQQQTWGYKPTQASL 25
Lambda K H
0.323 0.138 0.448
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4040519078
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40