bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
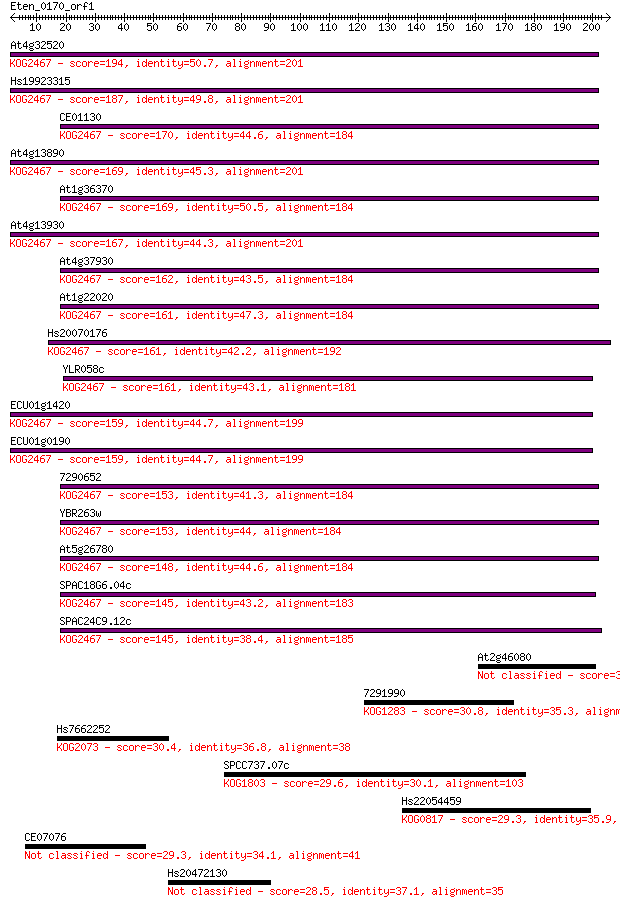
Query= Eten_0170_orf1
Length=205
Score E
Sequences producing significant alignments: (Bits) Value
At4g32520 194 8e-50
Hs19923315 187 2e-47
CE01130 170 2e-42
At4g13890 169 2e-42
At1g36370 169 5e-42
At4g13930 167 1e-41
At4g37930 162 5e-40
At1g22020 161 8e-40
Hs20070176 161 9e-40
YLR058c 161 1e-39
ECU01g1420 159 4e-39
ECU01g0190 159 4e-39
7290652 153 2e-37
YBR263w 153 3e-37
At5g26780 148 6e-36
SPAC18G6.04c 145 4e-35
SPAC24C9.12c 145 4e-35
At2g46080 32.3 0.74
7291990 30.8 1.8
Hs7662252 30.4 2.5
SPCC737.07c 29.6 4.2
Hs22054459 29.3 5.5
CE07076 29.3 6.2
Hs20472130 28.5 9.2
> At4g32520
Length=462
Score = 194 bits (494), Expect = 8e-50, Method: Compositional matrix adjust.
Identities = 102/211 (48%), Positives = 146/211 (69%), Gaps = 12/211 (5%)
Query 1 RSGIIFVNTRRVP-NGA---SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSR 56
R G+IF R+ P NG S +++AVFPGLQGGPH + I +A LK +PE++A+ +
Sbjct 253 RGGMIFF--RKDPINGVDLESAVNNAVFPGLQGGPHNHTIGGLAVCLKHAQSPEFKAYQK 310
Query 57 CVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSI 116
V+ NC+ LA L G +LV+GG+DNHL+L+DL+P G+ GA+++ I DMA+ITLNKNS+
Sbjct 311 RVVSNCRALANRLVELGFKLVSGGSDNHLVLVDLRPMGMDGARVEKILDMASITLNKNSV 370
Query 117 AGDSSG-IPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKS-GKKLAEFRK 174
GD S +P G+RIG+PAMTTRG ++F +VADFI+E V+I E ++ + G KL +F K
Sbjct 371 PGDKSALVPGGIRIGSPAMTTRGLSEKDFVVVADFIKEGVEITMEAKKAAPGSKLQDFNK 430
Query 175 HV-SPSHP---RISNLRDRVKALARSFPMPG 201
V SP P R+ +L++RV+ FP+PG
Sbjct 431 FVTSPEFPLKERVKSLKERVETFTSRFPIPG 461
> Hs19923315
Length=504
Score = 187 bits (474), Expect = 2e-47, Method: Compositional matrix adjust.
Identities = 100/216 (46%), Positives = 140/216 (64%), Gaps = 16/216 (7%)
Query 1 RSGIIFVNT----------RRVPNG-ASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
RSG+IF R +P RI+ AVFP LQGGPH + IAAVA LKQ CTP
Sbjct 286 RSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTP 345
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
+R +S VL+N + +A L G LV+GGTDNHL+L+DL+P+GL GA+ + + ++ +I
Sbjct 346 MFREYSLQVLKNARAMADALLERGYSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSI 405
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
T NKN+ GD S I P G+R+G PA+T+R F ++F+ V DFI E V+I E++ K+ K
Sbjct 406 TANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDEGVNIGLEVKSKTA-K 464
Query 169 LAEFRKHV---SPSHPRISNLRDRVKALARSFPMPG 201
L +F+ + S + R++NLR RV+ AR+FPMPG
Sbjct 465 LQDFKSFLLKDSETSQRLANLRQRVEQFARAFPMPG 500
> CE01130
Length=484
Score = 170 bits (430), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 82/188 (43%), Positives = 123/188 (65%), Gaps = 4/188 (2%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+SAVFPGLQGGPH + IA +A L+Q + ++ + VL+N K LA ++ HG L
Sbjct 293 KINSAVFPGLQGGPHNHTIAGIAVALRQCLSEDFVQYGEQVLKNAKTLAERMKKHGYALA 352
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTDNHLLL+DL+P G+ GA+ + + D+A+I NKN+ GD S + P G+R+GTPA+T+
Sbjct 353 TGGTDNHLLLVDLRPIGVEGARAEHVLDLAHIACNKNTCPGDVSALRPGGIRLGTPALTS 412
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHP---RISNLRDRVKAL 193
RGF ++F+ V DFI E V I + ++GK L +F+ + P +++L RV+
Sbjct 413 RGFQEQDFEKVGDFIHEGVQIAKKYNAEAGKTLKDFKSFTETNEPFKKDVADLAKRVEEF 472
Query 194 ARSFPMPG 201
+ F +PG
Sbjct 473 STKFEIPG 480
> At4g13890
Length=470
Score = 169 bits (429), Expect = 2e-42, Method: Compositional matrix adjust.
Identities = 91/215 (42%), Positives = 132/215 (61%), Gaps = 17/215 (7%)
Query 1 RSGIIFVNTRRVPNGA-------------SRIDSAVFPGLQGGPHENHIAAVAHQLKQTC 47
R+G+IF R+ P A ++I+SAVFP LQ GPH N I A+A LKQ
Sbjct 250 RAGMIFY--RKGPKPAKKGQPEGEVYDFDAKINSAVFPALQSGPHNNKIGALAVALKQVM 307
Query 48 TPEWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMA 107
P ++ +++ V N LA L N G LVT GTDNHL+L DL+P GLTG K++ +C++
Sbjct 308 APSFKVYAKQVKANAACLASYLINKGYTLVTDGTDNHLILWDLRPLGLTGNKVEKVCELC 367
Query 108 NITLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSG 166
ITLN+N++ GD+S + P GVRIGTPAMT+RG ++F+ + +F+ AV I +IQE+ G
Sbjct 368 YITLNRNAVFGDTSFLAPGGVRIGTPAMTSRGLVEKDFEKIGEFLHRAVTITLDIQEQYG 427
Query 167 KKLAEFRKHVSPSHPRISNLRDRVKALARSFPMPG 201
K + +F K + ++ I ++ V+ F MPG
Sbjct 428 KVMKDFNKGLV-NNKEIDEIKADVEEFTYDFDMPG 461
> At1g36370
Length=578
Score = 169 bits (427), Expect = 5e-42, Method: Compositional matrix adjust.
Identities = 93/185 (50%), Positives = 124/185 (67%), Gaps = 2/185 (1%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ AVFP LQGGPH NHIAA+A LKQ TPE++A+ + + +N + LA L +LV
Sbjct 389 KINFAVFPSLQGGPHNNHIAALAIALKQVATPEYKAYIQQMKKNAQALAAALLRRKCRLV 448
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTDNHLLL DL P GLTG + +C+M +ITLNK +I GD+ I P GVRIGTPAMTT
Sbjct 449 TGGTDNHLLLWDLTPMGLTGKVYEKVCEMCHITLNKTAIFGDNGTISPGGVRIGTPAMTT 508
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHPRISNLRDRVKALARS 196
RG +F+ +ADF+ +A I +Q + GK EF K + ++ I+ LR+RV+A A
Sbjct 509 RGCIESDFETMADFLIKAAQITSALQREHGKSHKEFVKSLC-TNKDIAELRNRVEAFALQ 567
Query 197 FPMPG 201
+ MP
Sbjct 568 YEMPA 572
> At4g13930
Length=471
Score = 167 bits (423), Expect = 1e-41, Method: Compositional matrix adjust.
Identities = 89/213 (41%), Positives = 133/213 (62%), Gaps = 13/213 (6%)
Query 1 RSGIIFVNT------RRVPNGA-----SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTP 49
R+G+IF + P GA +I+ AVFP LQGGPH + I A+A LKQ TP
Sbjct 250 RAGMIFYRKGPKPPKKGQPEGAVYDFEDKINFAVFPALQGGPHNHQIGALAVALKQANTP 309
Query 50 EWRAFSRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANI 109
++ +++ V N L L + G Q+VT GT+NHL+L DL+P GLTG K++ +CD+ +I
Sbjct 310 GFKVYAKQVKANAVALGNYLMSKGYQIVTNGTENHLVLWDLRPLGLTGNKVEKLCDLCSI 369
Query 110 TLNKNSIAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK 168
TLNKN++ GDSS + P GVRIG PAMT+RG ++F+ + +F+ AV + +IQ+ GK
Sbjct 370 TLNKNAVFGDSSALAPGGVRIGAPAMTSRGLVEKDFEQIGEFLSRAVTLTLDIQKTYGKL 429
Query 169 LAEFRKHVSPSHPRISNLRDRVKALARSFPMPG 201
L +F K + ++ + L+ V+ + S+ MPG
Sbjct 430 LKDFNKGLV-NNKDLDQLKADVEKFSASYEMPG 461
> At4g37930
Length=517
Score = 162 bits (410), Expect = 5e-40, Method: Compositional matrix adjust.
Identities = 80/189 (42%), Positives = 122/189 (64%), Gaps = 5/189 (2%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ AVFPGLQGGPH + I +A LKQ T E++A+ VL N A+ L G +LV
Sbjct 319 KINQAVFPGLQGGPHNHTITGLAVALKQATTSEYKAYQEQVLSNSAKFAQTLMERGYELV 378
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGD-SSGIPTGVRIGTPAMTT 136
+GGTDNHL+L++LKP+G+ G++++ + + +I NKN++ GD S+ +P G+R+GTPA+T+
Sbjct 379 SGGTDNHLVLVNLKPKGIDGSRVEKVLEAVHIASNKNTVPGDVSAMVPGGIRMGTPALTS 438
Query 137 RGFGAEEFKLVADFIREAVDICCEIQ-EKSGKKLAEFRKHVSPS---HPRISNLRDRVKA 192
RGF E+F VA++ +AV I +++ E G KL +F + S I+ LR V+
Sbjct 439 RGFVEEDFAKVAEYFDKAVTIALKVKSEAQGTKLKDFVSAMESSSTIQSEIAKLRHEVEE 498
Query 193 LARSFPMPG 201
A+ FP G
Sbjct 499 FAKQFPTIG 507
> At1g22020
Length=599
Score = 161 bits (408), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 87/185 (47%), Positives = 119/185 (64%), Gaps = 2/185 (1%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ +VFP LQGGPH NHIAA+A LKQ +PE++ + R V +N K LA L + +L+
Sbjct 413 KINFSVFPSLQGGPHNNHIAALAIALKQAASPEYKLYMRQVKKNAKALASALISRKCKLI 472
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTDNHLLL DL P GLTG + +C+M +IT+NK +I ++ I P GVRIG+PAMT+
Sbjct 473 TGGTDNHLLLWDLTPLGLTGKVYEKVCEMCHITVNKVAIFSENGVISPGGVRIGSPAMTS 532
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRKHVSPSHPRISNLRDRVKALARS 196
RG EF+ +ADF+ A I Q + GK E K + I++LR++V+A A
Sbjct 533 RGCLEPEFETMADFLYRAAQIASAAQREHGKLQKEPLKSIYHCK-EIADLRNQVEAFATQ 591
Query 197 FPMPG 201
F MP
Sbjct 592 FAMPA 596
> Hs20070176
Length=483
Score = 161 bits (407), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 81/197 (41%), Positives = 126/197 (63%), Gaps = 5/197 (2%)
Query 14 NGASRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHG 73
N S I+SAVFPGLQGGPH + IA VA LKQ T E++ + V+ NC+ L+ L G
Sbjct 287 NLESLINSAVFPGLQGGPHNHAIAGVAVALKQAMTLEFKVYQHQVVANCRALSEALTELG 346
Query 74 LQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTP 132
++VTGG+DNHL+L+DL+ +G G + + + + +I NKN+ GD S + P+G+R+GTP
Sbjct 347 YKIVTGGSDNHLILVDLRSKGTDGGRAEKVLEACSIACNKNTCPGDRSALRPSGLRLGTP 406
Query 133 AMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKK--LAEFRKHVS--PSHPRISNLRD 188
A+T+RG ++F+ VA FI +++ +IQ +G + L EF++ ++ + LR+
Sbjct 407 ALTSRGLLEKDFQKVAHFIHRGIELTLQIQSDTGVRATLKEFKERLAGDKYQAAVQALRE 466
Query 189 RVKALARSFPMPGRPDI 205
V++ A FP+PG PD
Sbjct 467 EVESFASFFPLPGLPDF 483
> YLR058c
Length=469
Score = 161 bits (407), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 78/185 (42%), Positives = 118/185 (63%), Gaps = 4/185 (2%)
Query 19 IDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLVT 78
I+ +VFPG QGGPH + IAA+A LKQ TPE++ + VL+N K L E +N G +LV+
Sbjct 283 INFSVFPGHQGGPHNHTIAALATALKQAATPEFKEYQTQVLKNAKALESEFKNLGYRLVS 342
Query 79 GGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSG-IPTGVRIGTPAMTTR 137
GTD+H++L+ L+ +G+ GA+++ IC+ NI LNKNSI GD S +P GVRIG PAMTTR
Sbjct 343 NGTDSHMVLVSLREKGVDGARVEYICEKINIALNKNSIPGDKSALVPGGVRIGAPAMTTR 402
Query 138 GFGAEEFKLVADFIREAVDICCEIQE---KSGKKLAEFRKHVSPSHPRISNLRDRVKALA 194
G G E+F + +I +AV+ ++Q+ K +L +F+ V ++ + + A
Sbjct 403 GMGEEDFHRIVQYINKAVEFAQQVQQSLPKDACRLKDFKAKVDEGSDVLNTWKKEIYDWA 462
Query 195 RSFPM 199
+P+
Sbjct 463 GEYPL 467
> ECU01g1420
Length=460
Score = 159 bits (401), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 89/211 (42%), Positives = 129/211 (61%), Gaps = 14/211 (6%)
Query 1 RSGIIFVNTRRVPNGAS-----RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFS 55
R +IF NG + RI+ AVFP LQGGPH + IA +A L TPE+ ++
Sbjct 250 RGALIFYRRAVTKNGETVDLDARINFAVFPMLQGGPHNHTIAGIASALLHAGTPEFAEYT 309
Query 56 RCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNS 115
R V+EN + L LQ+ GL ++TGGTDNH+LL+DL+ G+ GA ++ +CD I+LN+N+
Sbjct 310 RRVVENSRELCSRLQSLGLDILTGGTDNHMLLVDLRSTGVDGAAVEHMCDALGISLNRNA 369
Query 116 IAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRK 174
I G+SS + P+G+R+GT A+T RGFG EE + V D I V +C E+ G+K+++
Sbjct 370 IVGNSSPLSPSGIRVGTYAVTARGFGPEEMREVGDIIGGVVKLCREM--TGGRKMSKADL 427
Query 175 HVSPSHPRISN------LRDRVKALARSFPM 199
H S R+ LR RV ALA ++P+
Sbjct 428 HRVTSDARVMGSEQVLVLRRRVCALAEAYPI 458
> ECU01g0190
Length=460
Score = 159 bits (401), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 89/211 (42%), Positives = 129/211 (61%), Gaps = 14/211 (6%)
Query 1 RSGIIFVNTRRVPNGAS-----RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFS 55
R +IF NG + RI+ AVFP LQGGPH + IA +A L TPE+ ++
Sbjct 250 RGALIFYRRAVTKNGETVDLDARINFAVFPMLQGGPHNHTIAGIASALLHAGTPEFAEYT 309
Query 56 RCVLENCKVLARELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNS 115
R V+EN + L LQ+ GL ++TGGTDNH+LL+DL+ G+ GA ++ +CD I+LN+N+
Sbjct 310 RRVVENSRELCSRLQSLGLDILTGGTDNHMLLVDLRSTGVDGAAVEHMCDALGISLNRNA 369
Query 116 IAGDSSGI-PTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEFRK 174
I G+SS + P+G+R+GT A+T RGFG EE + V D I V +C E+ G+K+++
Sbjct 370 IVGNSSPLSPSGIRVGTYAVTARGFGPEEMREVGDIIGGVVKLCREM--TGGRKMSKADL 427
Query 175 HVSPSHPRISN------LRDRVKALARSFPM 199
H S R+ LR RV ALA ++P+
Sbjct 428 HRVTSDARVMGSEQVLVLRRRVCALAEAYPI 458
> 7290652
Length=537
Score = 153 bits (387), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 76/189 (40%), Positives = 118/189 (62%), Gaps = 5/189 (2%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
RI+ AVFP LQGGPH N +A +A KQ +PE++A+ VL+N K L L + G Q+
Sbjct 345 RINQAVFPSLQGGPHNNAVAGIATAFKQAKSPEFKAYQTQVLKNAKALCDGLISRGYQVA 404
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
TGGTD HL+L+D++ GLTGAK + I + I NKN++ GD S + P+G+R+GTPA+TT
Sbjct 405 TGGTDVHLVLVDVRKAGLTGAKAEYILEEVGIACNKNTVPGDKSAMNPSGIRLGTPALTT 464
Query 137 RGFGAEEFKLVADFIREAVDICCEIQEKSGK-KLAEFRKHVSPS---HPRISNLRDRVKA 192
RG ++ + V FI A+ + + + +G K+ ++ K ++ + ++ +R V
Sbjct 465 RGLAEQDIEQVVAFIDAALKVGVQAAKLAGSPKITDYHKTLAENVELKAQVDEIRKNVAQ 524
Query 193 LARSFPMPG 201
+R FP+PG
Sbjct 525 FSRKFPLPG 533
> YBR263w
Length=565
Score = 153 bits (386), Expect = 3e-37, Method: Compositional matrix adjust.
Identities = 81/192 (42%), Positives = 121/192 (63%), Gaps = 9/192 (4%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ +VFPG QGGPH + I A+A LKQ +PE++ + + +++N K A+EL G +LV
Sbjct 373 KINFSVFPGHQGGPHNHTIGAMAVALKQAMSPEFKEYQQKIVDNSKWFAQELTKMGYKLV 432
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
+GGTDNHL+++DL + GA+++ I NI NKN+I GD S + P+G+RIGTPAMTT
Sbjct 433 SGGTDNHLIVIDLSGTQVDGARVETILSALNIAANKNTIPGDKSALFPSGLRIGTPAMTT 492
Query 137 RGFGAEEFKLVADFIREAVDICCEIQ--EKSGK-----KLAEFRKHVSPSHPRISNLRDR 189
RGFG EEF VA +I AV + ++ E + K +L EF+K + S ++ L
Sbjct 493 RGFGREEFSQVAKYIDSAVKLAENLKTLEPTTKLDARSRLNEFKKLCNES-SEVAALSGE 551
Query 190 VKALARSFPMPG 201
+ +P+PG
Sbjct 552 ISKWVGQYPVPG 563
> At5g26780
Length=532
Score = 148 bits (374), Expect = 6e-36, Method: Compositional matrix adjust.
Identities = 82/212 (38%), Positives = 124/212 (58%), Gaps = 28/212 (13%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLA----------- 66
RI+ AVFPGLQGGPH + I +A LKQ TPE++A+ VL NC A
Sbjct 311 RINQAVFPGLQGGPHNHTITGLAVALKQARTPEYKAYQDQVLRNCSKFAELDIRPTVIIS 370
Query 67 -----RELQNHGLQLVTGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGD-S 120
+ L G LV+GGTDNHL+L++LK +G+ G++++ + ++ +I NKN++ GD S
Sbjct 371 YGLSMQTLLAKGYDLVSGGTDNHLVLVNLKNKGIDGSRVEKVLELVHIAANKNTVPGDVS 430
Query 121 SGIPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKS--------GKKLAEF 172
+ +P G+R+GTPA+T+RGF E+F VA++ AV I +I+ +S G KL +F
Sbjct 431 AMVPGGIRMGTPALTSRGFIEEDFAKVAEYFDLAVKIALKIKAESQGIYKKSFGTKLKDF 490
Query 173 RKHVSPS---HPRISNLRDRVKALARSFPMPG 201
+ + +S LR+ V+ A+ FP G
Sbjct 491 VATMQSNEKLQSEMSKLREMVEEYAKQFPTIG 522
> SPAC18G6.04c
Length=472
Score = 145 bits (367), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 79/190 (41%), Positives = 115/190 (60%), Gaps = 7/190 (3%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ +VFPG QGGPH + I A+A L Q TPE+ + + VL N K +A G +LV
Sbjct 282 KINFSVFPGHQGGPHNHTITALAVALGQAKTPEFYQYQKDVLSNAKAMANAFITRGYKLV 341
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSG-IPTGVRIGTPAMTT 136
+GGTD HL+L+DL +G+ GA+++ I ++ NI+ NKN++ GD S IP G+R+GTPA TT
Sbjct 342 SGGTDTHLVLVDLTDKGVDGARVERILELVNISANKNTVPGDKSALIPRGLRLGTPACTT 401
Query 137 RGFGAEEFKLVADFIREAVDICCEIQE---KSGK-KLAEFRKHV--SPSHPRISNLRDRV 190
RGF ++F+ V + I E V + +I E K GK K +F+ +V I+ L+ V
Sbjct 402 RGFDEKDFERVVELIDEVVSLTKKINEAALKEGKSKFRDFKAYVGDGSKFSEIAKLKKEV 461
Query 191 KALARSFPMP 200
A F P
Sbjct 462 ITWAGKFDFP 471
> SPAC24C9.12c
Length=467
Score = 145 bits (367), Expect = 4e-35, Method: Compositional matrix adjust.
Identities = 71/191 (37%), Positives = 114/191 (59%), Gaps = 6/191 (3%)
Query 18 RIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAFSRCVLENCKVLARELQNHGLQLV 77
+I+ +VFPG QGGPH + I A+A LKQ P ++ + V++N KV E + G +L
Sbjct 276 KINFSVFPGHQGGPHNHTITALAVALKQCQEPAYKEYQAQVVKNAKVCEEEFKKRGYKLA 335
Query 78 TGGTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGI-PTGVRIGTPAMTT 136
GTD+H++L+D+K +G+ GA+ + + ++ NI NKN++ D S P+G+R+GTPAMTT
Sbjct 336 ADGTDSHMVLVDVKSKGVDGARAERVLELINIVTNKNTVPSDKSAFSPSGIRVGTPAMTT 395
Query 137 RGFGAEEFKLVADFIREAVDICCEIQE---KSGKKLAEFRKHV--SPSHPRISNLRDRVK 191
RGF ++F V D+I A+ +Q+ K KL +F+ + +P + L+ V
Sbjct 396 RGFKEQDFVRVVDYIDRALTFAANLQKELPKDANKLKDFKAKLGEGEQYPELVQLQKEVA 455
Query 192 ALARSFPMPGR 202
A SFP+ +
Sbjct 456 EWASSFPLADK 466
> At2g46080
Length=347
Score = 32.3 bits (72), Expect = 0.74, Method: Compositional matrix adjust.
Identities = 16/47 (34%), Positives = 27/47 (57%), Gaps = 7/47 (14%)
Query 161 IQEKSGKK-------LAEFRKHVSPSHPRISNLRDRVKALARSFPMP 200
+Q SG+K L +R+HV+ ++PRI N R + +L +S +P
Sbjct 135 LQSDSGEKYLQARSSLDSWRQHVNTNNPRIENCRAVLDSLVKSLSLP 181
> 7291990
Length=446
Score = 30.8 bits (68), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 6/51 (11%)
Query 122 GIPTGVRIGTPAMTTRGFGAEEFKLVADFIREAVDICCEIQEKSGKKLAEF 172
GI TGV+ G + TT KL+ DF++ AVD+ E+ + K+ F
Sbjct 319 GINTGVKWGAQSGTTFT------KLMGDFMKPAVDVVGELLNNTTVKVGVF 363
> Hs7662252
Length=927
Score = 30.4 bits (67), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 14/38 (36%), Positives = 20/38 (52%), Gaps = 0/38 (0%)
Query 17 SRIDSAVFPGLQGGPHENHIAAVAHQLKQTCTPEWRAF 54
+RI +AV L+ GP + HI+ V L C W +F
Sbjct 481 TRIANAVVQNLERGPVQTHISEVIRGLPADCRGRWESF 518
> SPCC737.07c
Length=660
Score = 29.6 bits (65), Expect = 4.2, Method: Composition-based stats.
Identities = 31/116 (26%), Positives = 53/116 (45%), Gaps = 14/116 (12%)
Query 74 LQLVTG--GTDNHLLLLDLKPQGLTGAKLQLICDMANITLNKNSIAGDSSGIPTGVRIGT 131
L L+ G GT L+++ Q + K L+C +N+ ++ SSGIP VR+G
Sbjct 227 LSLIHGPPGTGKTHTLVEIIQQLVLRNKRILVCGASNLAVDNIVDRLSSSGIPM-VRLGH 285
Query 132 PAMTTRGFGAEEFKLVA------DFIR---EAVDICCE--IQEKSGKKLAEFRKHV 176
PA +++ D IR E +D+C + K+G++ E K++
Sbjct 286 PARLLPSILDHSLDVLSRTGDNGDVIRGISEDIDVCLSKITKTKNGRERREIYKNI 341
> Hs22054459
Length=344
Score = 29.3 bits (64), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 23/65 (35%), Positives = 31/65 (47%), Gaps = 3/65 (4%)
Query 135 TTRGFGAEEFKLVAD-FIREAVDICCEIQEKSGKKLAEFRKHVSPSHPRISNLRDRVKAL 193
+ + F A FK V F+ V C+ K GK L+ F H + +S L D +KAL
Sbjct 73 SEKKFIAGLFKFVEKMFVGNRVG--CKSFGKKGKTLSLFVLHARLAVANVSGLDDSLKAL 130
Query 194 ARSFP 198
R FP
Sbjct 131 PRYFP 135
> CE07076
Length=230
Score = 29.3 bits (64), Expect = 6.2, Method: Compositional matrix adjust.
Identities = 14/41 (34%), Positives = 24/41 (58%), Gaps = 5/41 (12%)
Query 6 FVNTRRVPNGASRIDSAVFPGLQGGPHENHIAAVAHQLKQT 46
FVN R++ G + +D +FP L +H + A+Q+K+T
Sbjct 76 FVNGRKISAGLTDVDGYIFPCL-----ASHCPSAANQVKET 111
> Hs20472130
Length=149
Score = 28.5 bits (62), Expect = 9.2, Method: Compositional matrix adjust.
Identities = 13/35 (37%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 55 SRCVLENCKVLARELQNHGLQLVTGGTDNHLLLLD 89
SRC+L++C L R+ NH + T + LLL++
Sbjct 113 SRCILDHCNWLLRDEDNHRSGEIGAYTTSFLLLVE 147
Lambda K H
0.321 0.137 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3658712052
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40