bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0163_orf1
Length=110
Score E
Sequences producing significant alignments: (Bits) Value
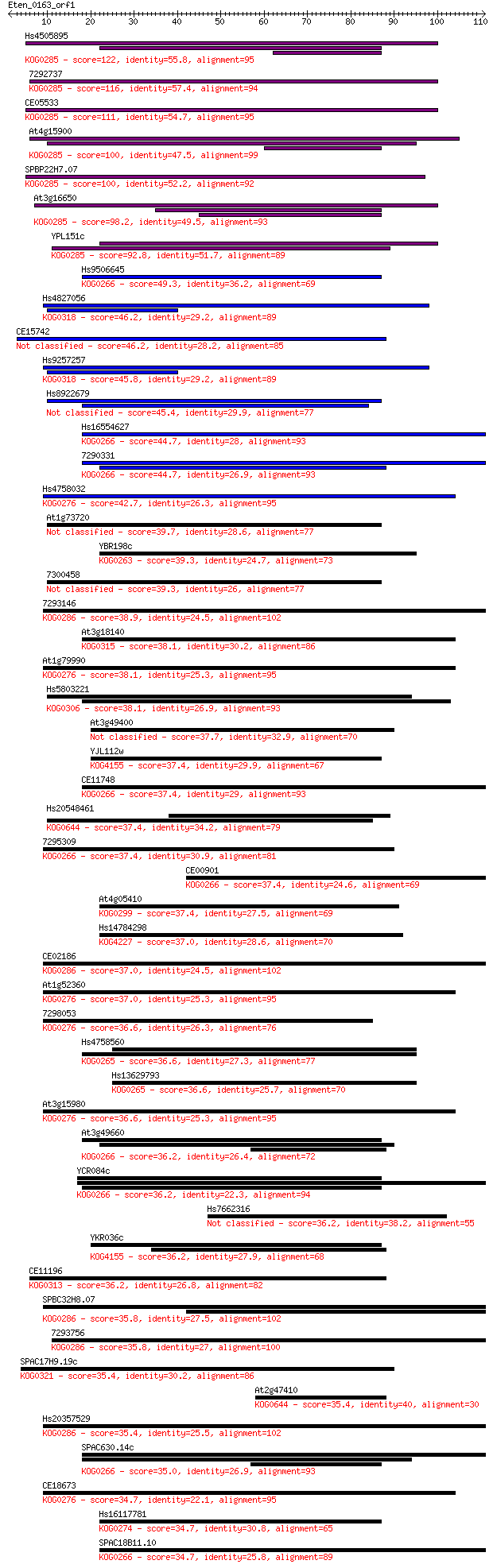
Hs4505895 122 2e-28
7292737 116 8e-27
CE05533 111 3e-25
At4g15900 100 7e-22
SPBP22H7.07 100 9e-22
At3g16650 98.2 3e-21
YPL151c 92.8 1e-19
Hs9506645 49.3 2e-06
Hs4827056 46.2 1e-05
CE15742 46.2 2e-05
Hs9257257 45.8 2e-05
Hs8922679 45.4 3e-05
Hs16554627 44.7 5e-05
7290331 44.7 5e-05
Hs4758032 42.7 1e-04
At1g73720 39.7 0.001
YBR198c 39.3 0.002
7300458 39.3 0.002
7293146 38.9 0.003
At3g18140 38.1 0.004
At1g79990 38.1 0.004
Hs5803221 38.1 0.004
At3g49400 37.7 0.005
YJL112w 37.4 0.006
CE11748 37.4 0.006
Hs20548461 37.4 0.006
7295309 37.4 0.007
CE00901 37.4 0.007
At4g05410 37.4 0.008
Hs14784298 37.0 0.008
CE02186 37.0 0.008
At1g52360 37.0 0.009
7298053 36.6 0.011
Hs4758560 36.6 0.012
Hs13629793 36.6 0.012
At3g15980 36.6 0.013
At3g49660 36.2 0.014
YCR084c 36.2 0.015
Hs7662316 36.2 0.016
YKR036c 36.2 0.016
CE11196 36.2 0.017
SPBC32H8.07 35.8 0.018
7293756 35.8 0.022
SPAC17H9.19c 35.4 0.025
At2g47410 35.4 0.028
Hs20357529 35.4 0.028
SPAC630.14c 35.0 0.037
CE18673 34.7 0.039
Hs16117781 34.7 0.040
SPAC18B11.10 34.7 0.040
> Hs4505895
Length=514
Score = 122 bits (305), Expect = 2e-28, Method: Composition-based stats.
Identities = 53/95 (55%), Positives = 72/95 (75%), Gaps = 3/95 (3%)
Query 5 NSIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCA 64
N+I N +G LV G++NG +H WDWR+GY +Q + + VQPGSL+SE+GIF CA
Sbjct 413 NAIINTLTVNSDG---VLVSGADNGTMHLWDWRTGYNFQRVHAAVQPGSLDSESGIFACA 469
Query 65 FDKSETRLITGECDKTIKIWKIDEEATEETHPISW 99
FD+SE+RL+T E DKTIK+++ D+ ATEETHP+SW
Sbjct 470 FDQSESRLLTAEADKTIKVYREDDTATEETHPVSW 504
Score = 28.5 bits (62), Expect = 2.8, Method: Composition-based stats.
Identities = 17/65 (26%), Positives = 26/65 (40%), Gaps = 8/65 (12%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
LV S + WD R+ TL N + +E ++ITG D TI
Sbjct 303 LVTCSRDSTARIWDVRTKASVHTLSGHT--------NAVATVRCQAAEPQIITGSHDTTI 354
Query 82 KIWKI 86
++W +
Sbjct 355 RLWDL 359
Score = 27.3 bits (59), Expect = 7.7, Method: Composition-based stats.
Identities = 10/25 (40%), Positives = 14/25 (56%), Gaps = 0/25 (0%)
Query 62 CCAFDKSETRLITGECDKTIKIWKI 86
C A + +TG D+TIKIW +
Sbjct 209 CIAVEPGNQWFVTGSADRTIKIWDL 233
> 7292737
Length=482
Score = 116 bits (291), Expect = 8e-27, Method: Composition-based stats.
Identities = 54/94 (57%), Positives = 69/94 (73%), Gaps = 3/94 (3%)
Query 6 SIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAF 65
SI NC A EG LV G +NG + FWDWR+GY +Q Q+ VQPGS++SE GIF F
Sbjct 382 SIVNCMAANSEG---VLVSGGDNGTMFFWDWRTGYNFQRFQAPVQPGSMDSEAGIFAMCF 438
Query 66 DKSETRLITGECDKTIKIWKIDEEATEETHPISW 99
D+S +RLIT E DKTIK++K D+EA+EE+HPI+W
Sbjct 439 DQSGSRLITAEADKTIKVYKEDDEASEESHPINW 472
> CE05533
Length=494
Score = 111 bits (278), Expect = 3e-25, Method: Composition-based stats.
Identities = 52/95 (54%), Positives = 70/95 (73%), Gaps = 3/95 (3%)
Query 5 NSIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCA 64
N+I N + D+G +V G++NG L FWDWRSG+ +Q +Q++ QPGS+ESE GI+
Sbjct 393 NAIINTLSSNDDG---VVVSGADNGSLCFWDWRSGFCFQKIQTKPQPGSIESEAGIYASC 449
Query 65 FDKSETRLITGECDKTIKIWKIDEEATEETHPISW 99
FDK+ RLIT E DKTIK++K D+EATEE+HPI W
Sbjct 450 FDKTGLRLITAEADKTIKMYKEDDEATEESHPIVW 484
> At4g15900
Length=486
Score = 100 bits (249), Expect = 7e-22, Method: Compositional matrix adjust.
Identities = 47/99 (47%), Positives = 68/99 (68%), Gaps = 3/99 (3%)
Query 6 SIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAF 65
+I N A ++G +V G +NG + FWDW+SG+ +Q ++ VQPGSLESE GI+ +
Sbjct 387 TIINAMAVNEDG---VMVTGGDNGSIWFWDWKSGHSFQQSETIVQPGSLESEAGIYAACY 443
Query 66 DKSETRLITGECDKTIKIWKIDEEATEETHPISWGHKRD 104
D + +RL+T E DKTIK+WK DE AT ETHPI++ ++
Sbjct 444 DNTGSRLVTCEADKTIKMWKEDENATPETHPINFKPPKE 482
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 21/86 (24%), Positives = 35/86 (40%), Gaps = 12/86 (13%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDK-S 68
C A D L+ G + WD R+ ++Q +L + C F + +
Sbjct 265 CLALHPTLD--VLLTGGRDSVCRVWDIRT---------KMQIFALSGHDNTVCSVFTRPT 313
Query 69 ETRLITGECDKTIKIWKIDEEATEET 94
+ +++TG D TIK W + T T
Sbjct 314 DPQVVTGSHDTTIKFWDLRYGKTMST 339
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 12/27 (44%), Positives = 15/27 (55%), Gaps = 0/27 (0%)
Query 60 IFCCAFDKSETRLITGECDKTIKIWKI 86
+ AFD S TG D+TIKIW +
Sbjct 179 VRSVAFDPSNEWFCTGSADRTIKIWDV 205
> SPBP22H7.07
Length=473
Score = 100 bits (248), Expect = 9e-22, Method: Compositional matrix adjust.
Identities = 48/92 (52%), Positives = 65/92 (70%), Gaps = 3/92 (3%)
Query 5 NSIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCA 64
N+I N + + + + G++NG + FWDW+SG+KYQ LQS VQPGSL+SE GIF +
Sbjct 372 NAIVNTLSINSD---NVMFSGADNGSMCFWDWKSGHKYQELQSVVQPGSLDSEAGIFASS 428
Query 65 FDKSETRLITGECDKTIKIWKIDEEATEETHP 96
FDK+ RLIT E DK++KI+K + AT ETHP
Sbjct 429 FDKTGLRLITCEADKSVKIYKQVDNATPETHP 460
> At3g16650
Length=477
Score = 98.2 bits (243), Expect = 3e-21, Method: Compositional matrix adjust.
Identities = 46/93 (49%), Positives = 66/93 (70%), Gaps = 3/93 (3%)
Query 7 IPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFD 66
I N A ++G +V G + G L FWDW+SG+ +Q ++ VQPGSLESE GI+ +D
Sbjct 379 IINAVAVNEDG---VMVTGGDKGGLWFWDWKSGHNFQRAETIVQPGSLESEAGIYAACYD 435
Query 67 KSETRLITGECDKTIKIWKIDEEATEETHPISW 99
++ +RL+T E DKTIK+WK DE+AT ETHP+++
Sbjct 436 QTGSRLVTCEGDKTIKMWKEDEDATPETHPLNF 468
Score = 28.5 bits (62), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 17/53 (32%), Positives = 25/53 (47%), Gaps = 9/53 (16%)
Query 35 DWRSGYK-YQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIWKI 86
+W + +K Y+ LQ + + AFD S TG D+TIKIW +
Sbjct 153 EWHAPWKNYRVLQGHL--------GWVRSVAFDPSNEWFCTGSADRTIKIWDV 197
Score = 28.1 bits (61), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 11/42 (26%), Positives = 24/42 (57%), Gaps = 0/42 (0%)
Query 45 LQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIWKI 86
+++++Q L ++ +F ++ ++ITG D TIK W +
Sbjct 281 IRTKMQIFVLPHDSDVFSVLARPTDPQVITGSHDSTIKFWDL 322
> YPL151c
Length=451
Score = 92.8 bits (229), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 43/79 (54%), Positives = 57/79 (72%), Gaps = 1/79 (1%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
L G +NG L F+D++SG+KYQ+L +R GSLE E + C FDK+ RLITGE DK+I
Sbjct 364 LFAGGDNGVLSFYDYKSGHKYQSLATREMVGSLEGERSVLCSTFDKTGLRLITGEADKSI 423
Query 82 KIWKIDEEATEETHP-ISW 99
KIWK DE AT+E+ P ++W
Sbjct 424 KIWKQDETATKESEPGLAW 442
Score = 28.1 bits (61), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 19/78 (24%), Positives = 29/78 (37%), Gaps = 8/78 (10%)
Query 11 CAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSET 70
C D D + GSN+ + WD +G TL V + A
Sbjct 144 CVAIDPVDNEWFITGSNDTTMKVWDLATGKLKTTLAGHVMT--------VRDVAVSDRHP 195
Query 71 RLITGECDKTIKIWKIDE 88
L + DKT+K W +++
Sbjct 196 YLFSVSEDKTVKCWDLEK 213
> Hs9506645
Length=330
Score = 49.3 bits (116), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 37/69 (53%), Gaps = 8/69 (11%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D S LV S++ L WD RSG +TL+ N +FCC F+ +I+G
Sbjct 94 DSSRLVSASDDKTLKLWDVRSGKCLKTLKGH--------SNYVFCCNFNPPSNLIISGSF 145
Query 78 DKTIKIWKI 86
D+T+KIW++
Sbjct 146 DETVKIWEV 154
> Hs4827056
Length=534
Score = 46.2 bits (108), Expect = 1e-05, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 7/89 (7%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC F +G+R S +GQ++ +D ++G K L GS + GI+ ++
Sbjct 193 NCVRFSPDGNR--FATASADGQIYIYDGKTGEKVCALG-----GSKAHDGGIYAISWSPD 245
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPI 97
T L++ DKT KIW + + T P+
Sbjct 246 STHLLSASGDKTSKIWDVSVNSVVSTFPM 274
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 9/30 (30%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSG 39
C G +S + GS++G +++WD +G
Sbjct 325 CLTVHKNGGKSYIYSGSHDGHINYWDSETG 354
> CE15742
Length=510
Score = 46.2 bits (108), Expect = 2e-05, Method: Composition-based stats.
Identities = 24/85 (28%), Positives = 41/85 (48%), Gaps = 2/85 (2%)
Query 3 SLNSIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFC 62
S S P F D + LV GS +G + W++ +G + L+ + Q + + + C
Sbjct 212 STKSYPESAVFSP--DANYLVSGSKDGFIEVWNYMNGKLRKDLKYQAQDNLMMMDAAVRC 269
Query 63 CAFDKSETRLITGECDKTIKIWKID 87
+F + L TG D IK+WK++
Sbjct 270 ISFSRDSEMLATGSIDGKIKVWKVE 294
> Hs9257257
Length=606
Score = 45.8 bits (107), Expect = 2e-05, Method: Composition-based stats.
Identities = 26/89 (29%), Positives = 43/89 (48%), Gaps = 7/89 (7%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC F +G+R S +GQ++ +D ++G K L GS + GI+ ++
Sbjct 193 NCVRFSPDGNR--FATASADGQIYIYDGKTGEKVCALG-----GSKAHDGGIYAISWSPD 245
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPI 97
T L++ DKT KIW + + T P+
Sbjct 246 STHLLSASGDKTSKIWDVSVNSVVSTFPM 274
Score = 26.9 bits (58), Expect = 8.8, Method: Composition-based stats.
Identities = 9/30 (30%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSG 39
C G +S + GS++G +++WD +G
Sbjct 325 CLTVHKNGGKSYIYSGSHDGHINYWDSETG 354
> Hs8922679
Length=513
Score = 45.4 bits (106), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 23/77 (29%), Positives = 38/77 (49%), Gaps = 2/77 (2%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSE 69
C F +G LV GS +G + W++ +G + L+ + Q + ++ + C F +
Sbjct 219 CARFSPDGQY--LVTGSVDGFIEVWNFTTGKIRKDLKYQAQDNFMMMDDAVLCMCFSRDT 276
Query 70 TRLITGECDKTIKIWKI 86
L TG D IK+WKI
Sbjct 277 EMLATGAQDGKIKVWKI 293
Score = 31.2 bits (69), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 18/66 (27%), Positives = 34/66 (51%), Gaps = 7/66 (10%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D L G+ +G++ W +SG Q L+ + S G+ C +F K +++++
Sbjct 275 DTEMLATGAQDGKIKVWKIQSG---QCLRRFERAHS----KGVTCLSFSKDSSQILSASF 327
Query 78 DKTIKI 83
D+TI+I
Sbjct 328 DQTIRI 333
> Hs16554627
Length=334
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 26/93 (27%), Positives = 43/93 (46%), Gaps = 11/93 (11%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D + LV S++ L WD SG +TL+ N +FCC F+ +++G
Sbjct 98 DSNLLVSASDDKTLKIWDVSSGKCLKTLKGH--------SNYVFCCNFNPQSNLIVSGSF 149
Query 78 DKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
D++++IW + +T P H +AV F
Sbjct 150 DESVRIWDVKTGKCLKTLP---AHSDPVSAVHF 179
> 7290331
Length=361
Score = 44.7 bits (104), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 25/93 (26%), Positives = 43/93 (46%), Gaps = 11/93 (11%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D LV GS++ L W+ +G +TL+ N +FCC F+ +++G
Sbjct 125 DSRLLVSGSDDKTLKVWELSTGKSLKTLKGH--------SNYVFCCNFNPQSNLIVSGSF 176
Query 78 DKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
D++++IW + +T P H +AV F
Sbjct 177 DESVRIWDVRTGKCLKTLP---AHSDPVSAVHF 206
Score = 31.2 bits (69), Expect = 0.44, Method: Compositional matrix adjust.
Identities = 20/68 (29%), Positives = 32/68 (47%), Gaps = 10/68 (14%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITG--ECDK 79
+V GS + ++ W+ +S Q LQ + + C A +E + + E DK
Sbjct 301 IVSGSEDNMVYIWNLQSKEVVQKLQGHT--------DTVLCTACHPTENIIASAALENDK 352
Query 80 TIKIWKID 87
TIK+WK D
Sbjct 353 TIKLWKSD 360
> Hs4758032
Length=906
Score = 42.7 bits (99), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 25/95 (26%), Positives = 45/95 (47%), Gaps = 10/95 (10%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC + GD+ L+ G+++ + WD+++ QTL+ Q + C +F
Sbjct 189 NCIDYYSGGDKPYLISGADDRLVKIWDYQNKTCVQTLEGHAQ--------NVSCASFHPE 240
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKR 103
+ITG D T++IW E T +++G +R
Sbjct 241 LPIIITGSEDGTVRIWHSSTYRLEST--LNYGMER 273
> At1g73720
Length=522
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSE 69
C F +G L S +G + WD+ SG + LQ + + ++ + C F +
Sbjct 229 CARFSPDGQ--FLASSSVDGFIEVWDYISGKLKKDLQYQADESFMMHDDPVLCIDFSRDS 286
Query 70 TRLITGECDKTIKIWKI 86
L +G D IKIW+I
Sbjct 287 EMLASGSQDGKIKIWRI 303
> YBR198c
Length=798
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 18/73 (24%), Positives = 37/73 (50%), Gaps = 7/73 (9%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
L GS +G ++ WD +G + + ++ +N I+ ++ K LI+G D T+
Sbjct 666 LSTGSEDGIINVWDIGTGKRLKQMRG-------HGKNAIYSLSYSKEGNVLISGGADHTV 718
Query 82 KIWKIDEEATEET 94
++W + + TE +
Sbjct 719 RVWDLKKATTEPS 731
> 7300458
Length=486
Score = 39.3 bits (90), Expect = 0.002, Method: Composition-based stats.
Identities = 20/77 (25%), Positives = 36/77 (46%), Gaps = 2/77 (2%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSE 69
C F +G L+ GS +G L W++ +G + L+ + Q + E + F +
Sbjct 196 CAQFSPDG--QYLITGSVDGFLEVWNFTTGKVRKDLKYQAQDQFMMMEQAVLALNFSRDS 253
Query 70 TRLITGECDKTIKIWKI 86
+ +G D IK+W+I
Sbjct 254 EMVASGAQDGQIKVWRI 270
> 7293146
Length=340
Score = 38.9 bits (89), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 42/102 (41%), Gaps = 14/102 (13%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
+CC F D+ + +V S + WD +G + + L + +
Sbjct 147 SCCRFLDD---NQIVTSSGDMSCGLWDIETGLQVTSF--------LGHTGDVMALSLAPQ 195
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
++G CD + K+W I E ++T P GH+ D AV F
Sbjct 196 CKTFVSGACDASAKLWDIREGVCKQTFP---GHESDINAVTF 234
> At3g18140
Length=305
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 26/87 (29%), Positives = 39/87 (44%), Gaps = 6/87 (6%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENG-IFCCAFDKSETRLITGE 76
D + +V +N G + W G QT+ L++ NG I C + L T
Sbjct 173 DGTMVVAANNRGTCYVWRLLRGK--QTMTEFEPLHKLQAHNGHILKCLLSPANKYLATAS 230
Query 77 CDKTIKIWKIDEEATEETHPISWGHKR 103
DKT+KIW +D E+ + GH+R
Sbjct 231 SDKTVKIWNVDGFKLEK---VLTGHQR 254
> At1g79990
Length=913
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 43/95 (45%), Gaps = 10/95 (10%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC + GD+ L+ GS++ WD+++ QTL+ + + +F
Sbjct 182 NCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLEGHT--------HNVSAVSFHPE 233
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKR 103
+ITG D T++IW E T +++G +R
Sbjct 234 LPIIITGSEDGTVRIWHATTYRLENT--LNYGLER 266
> Hs5803221
Length=943
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 22/88 (25%), Positives = 42/88 (47%), Gaps = 13/88 (14%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSE 69
C+F GDR +V+G+ G+L +D SG +T+ + + ++ + +
Sbjct 458 LCSFFVPGDRQ-VVIGTKTGKLQLYDLASGNLLETIDAH--------DGALWSMSLSPDQ 508
Query 70 TRLITGECDKTIKIWKI----DEEATEE 93
+TG DK++K W DE +T++
Sbjct 509 RGFVTGGADKSVKFWDFELVKDENSTQK 536
Score = 33.9 bits (76), Expect = 0.086, Method: Compositional matrix adjust.
Identities = 21/88 (23%), Positives = 40/88 (45%), Gaps = 6/88 (6%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRV---QPGSLESENGIFCCAFDKSETRLIT 74
D+ V G + + FWD+ + Q R+ Q +L+ + + C ++ ++ L
Sbjct 507 DQRGFVTGGADKSVKFWDFELVKDENSTQKRLSVKQTRTLQLDEDVLCVSYSPNQKLLAV 566
Query 75 GECDKTIKIWKIDEEATEETHPISWGHK 102
D T+KI+ +D T + +GHK
Sbjct 567 SLLDCTVKIFYVD---TLKFFLSLYGHK 591
> At3g49400
Length=719
Score = 37.7 bits (86), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 13/83 (15%)
Query 20 SSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQP-----GSLESEN--------GIFCCAFD 66
S L +GS +G + W + Y +S V P +++ + GIF C
Sbjct 306 SLLAIGSKSGSVSIWKVHAPECYHIERSNVSPMVELTAIVQTHSSWVSTMSWGIFGCDSS 365
Query 67 KSETRLITGECDKTIKIWKIDEE 89
+ L+TG CD ++KIW ++E
Sbjct 366 NPQVVLVTGSCDGSVKIWMSNKE 388
> YJL112w
Length=714
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 20/67 (29%), Positives = 34/67 (50%), Gaps = 10/67 (14%)
Query 20 SSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDK 79
++L G+ +G + WD RSG +TL+ + I FD + L+TG D+
Sbjct 577 AALATGTKDGVVRLWDLRSGKVIRTLKGHT--------DAITSLKFDSA--CLVTGSYDR 626
Query 80 TIKIWKI 86
T++IW +
Sbjct 627 TVRIWDL 633
> CE11748
Length=395
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 27/93 (29%), Positives = 44/93 (47%), Gaps = 11/93 (11%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D +V S++ + +D SG +TL+ N +FCC F+ S T + +G
Sbjct 159 DSKLIVSCSDDKLVKVFDVSSGRCVKTLKGHT--------NYVFCCCFNPSGTLIASGSF 210
Query 78 DKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
D+TI+IW T + P GH+ ++V F
Sbjct 211 DETIRIWCARNGNTIFSIP---GHEDPVSSVCF 240
> Hs20548461
Length=736
Score = 37.4 bits (85), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 4/51 (7%)
Query 38 SGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIWKIDE 88
S Y++ + R+ G L S ++C AFD+S R+ TG D +KIW D+
Sbjct 196 SAYQHIKMHKRIL-GHLSS---VYCVAFDRSGRRIFTGSDDCLVKIWATDD 242
Score = 27.3 bits (59), Expect = 7.7, Method: Compositional matrix adjust.
Identities = 21/75 (28%), Positives = 31/75 (41%), Gaps = 10/75 (13%)
Query 10 CCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSE 69
C AF G R + GS++ + W G TL+ S E I A +
Sbjct 216 CVAFDRSGRR--IFTGSDDCLVKIWATDDGRLLATLRGH----SAE----ISDMAVNYEN 265
Query 70 TRLITGECDKTIKIW 84
T + G CDK +++W
Sbjct 266 TLIAAGSCDKVVRVW 280
> 7295309
Length=742
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 25/81 (30%), Positives = 39/81 (48%), Gaps = 10/81 (12%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC A + GD + VGS+ Q+ WD+ SG + V GS E + + CA+
Sbjct 584 NCLAINETGDYF-ITVGSDL-QVKLWDYNSG-------AVVGIGS-EHASSVISCAYSPC 633
Query 69 ETRLITGECDKTIKIWKIDEE 89
T +TG D ++ IW + +
Sbjct 634 MTMFVTGSTDGSLIIWDVPRD 654
> CE00901
Length=376
Score = 37.4 bits (85), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 17/69 (24%), Positives = 35/69 (50%), Gaps = 3/69 (4%)
Query 42 YQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIWKIDEEATEETHPISWGH 101
++ + SR+ N +FCC F+ + +++G D++++IW + +T P H
Sbjct 156 FEIVTSRMTKTLKGHNNYVFCCNFNPQSSLVVSGSFDESVRIWDVKTGMCIKTLP---AH 212
Query 102 KRDRAAVAF 110
+AV+F
Sbjct 213 SDPVSAVSF 221
> At4g05410
Length=504
Score = 37.4 bits (85), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 19/69 (27%), Positives = 32/69 (46%), Gaps = 8/69 (11%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
L G + +H WD R+ Q PG N + C F + L +G D+T+
Sbjct 237 LATGGVDRHVHIWDVRTREHVQAF-----PGH---RNTVSCLCFRYGTSELYSGSFDRTV 288
Query 82 KIWKIDEEA 90
K+W ++++A
Sbjct 289 KVWNVEDKA 297
> Hs14784298
Length=942
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 20/72 (27%), Positives = 38/72 (52%), Gaps = 7/72 (9%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESEN--GIFCCAFDKSETRLITGECDK 79
LV G ++ ++ W + Q + SRV+P L+ E+ IFC AF+ T++ +G D+
Sbjct 69 LVSGGDDRRVLLW-----HMEQAIHSRVKPIQLKGEHHSNIFCLAFNSGNTKVFSGGNDE 123
Query 80 TIKIWKIDEEAT 91
+ + ++ T
Sbjct 124 QVILHDVESSET 135
> CE02186
Length=340
Score = 37.0 bits (84), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 25/102 (24%), Positives = 41/102 (40%), Gaps = 14/102 (13%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
+CC F D+ + +V S + WD +G + + +
Sbjct 147 SCCRFLDD---NQIVTSSGDMTCALWDIETGQQCTAFTGHT--------GDVMSLSLSPD 195
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
I+G CD + K+W I + ++T P GH+ D AVAF
Sbjct 196 FRTFISGACDASAKLWDIRDGMCKQTFP---GHESDINAVAF 234
> At1g52360
Length=926
Score = 37.0 bits (84), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 42/95 (44%), Gaps = 10/95 (10%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC + GD+ L+ GS++ WD+++ QTL+ + + F
Sbjct 189 NCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLEGHT--------HNVSAVCFHPE 240
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKR 103
+ITG D T++IW E T +++G +R
Sbjct 241 LPIIITGSEDGTVRIWHATTYRLENT--LNYGLER 273
> 7298053
Length=860
Score = 36.6 bits (83), Expect = 0.011, Method: Composition-based stats.
Identities = 20/76 (26%), Positives = 35/76 (46%), Gaps = 8/76 (10%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC + GD+ L+ G+++ + WD+++ QTL+ Q I F
Sbjct 160 NCVDYYHGGDKPYLISGADDRLVKIWDYQNKTCVQTLEGHAQ--------NISAVCFHPE 211
Query 69 ETRLITGECDKTIKIW 84
++TG D T++IW
Sbjct 212 LPIVLTGSEDGTVRIW 227
> Hs4758560
Length=357
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 31/70 (44%), Gaps = 9/70 (12%)
Query 25 GSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIW 84
GS++G + WD R QT Q+ Q + F+ + ++I+G D IK+W
Sbjct 170 GSDDGTVKLWDIRKKAAIQTFQNTYQ---------VLAVTFNDTSDQIISGGIDNDIKVW 220
Query 85 KIDEEATEET 94
+ + T
Sbjct 221 DLRQNKLTYT 230
Score = 28.5 bits (62), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 22/77 (28%), Positives = 34/77 (44%), Gaps = 7/77 (9%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D S L S + + WD +G + + L+ G N C KS + TG
Sbjct 120 DGSMLFSASTDKTVAVWDSETGERVKRLK-----GHTSFVNS--CYPARKSPQLVCTGSD 172
Query 78 DKTIKIWKIDEEATEET 94
D T+K+W I ++A +T
Sbjct 173 DGTVKLWDIRKKAAIQT 189
> Hs13629793
Length=357
Score = 36.6 bits (83), Expect = 0.012, Method: Compositional matrix adjust.
Identities = 18/70 (25%), Positives = 31/70 (44%), Gaps = 9/70 (12%)
Query 25 GSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIW 84
GS++G + WD R QT Q+ Q + F+ + ++I+G D IK+W
Sbjct 170 GSDDGTVKLWDIRKKAAIQTFQNTYQ---------VLAVTFNDTSDQIISGGIDNDIKVW 220
Query 85 KIDEEATEET 94
+ + T
Sbjct 221 DLRQNKLTYT 230
> At3g15980
Length=921
Score = 36.6 bits (83), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 24/95 (25%), Positives = 41/95 (43%), Gaps = 10/95 (10%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC + GD+ L+ GS++ WD+++ QTL + + F
Sbjct 192 NCVDYFTGGDKPYLITGSDDHTAKVWDYQTKSCVQTLDGHT--------HNVSAVCFHPE 243
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKR 103
+ITG D T++IW E T +++G +R
Sbjct 244 LPIIITGSEDGTVRIWHATTYRLENT--LNYGLER 276
> At3g49660
Length=317
Score = 36.2 bits (82), Expect = 0.014, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 8/69 (11%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D +V S++ L WD +G +TL + N FC F+ +++G
Sbjct 82 DARFIVSASDDKTLKLWDVETGSLIKTL--------IGHTNYAFCVNFNPQSNMIVSGSF 133
Query 78 DKTIKIWKI 86
D+T++IW +
Sbjct 134 DETVRIWDV 142
Score = 31.2 bits (69), Expect = 0.54, Method: Compositional matrix adjust.
Identities = 19/68 (27%), Positives = 32/68 (47%), Gaps = 8/68 (11%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
+V GS + +H W+ S Q L+ G E+ + C +E + +G DKT+
Sbjct 258 IVSGSEDNCVHMWELNSKKLLQKLE-----GHTETVMNVAC---HPTENLIASGSLDKTV 309
Query 82 KIWKIDEE 89
+IW +E
Sbjct 310 RIWTQKKE 317
Score = 27.3 bits (59), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 0/31 (0%)
Query 57 ENGIFCCAFDKSETRLITGECDKTIKIWKID 87
ENGI AF +++ DKT+K+W ++
Sbjct 71 ENGISDVAFSSDARFIVSASDDKTLKLWDVE 101
> YCR084c
Length=713
Score = 36.2 bits (82), Expect = 0.015, Method: Composition-based stats.
Identities = 15/70 (21%), Positives = 36/70 (51%), Gaps = 1/70 (1%)
Query 17 GDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGE 76
GD + GS + + WD +G+ + L S + G+ ++ ++ F + +++G
Sbjct 537 GDGKYIAAGSLDRAVRVWDSETGFLVERLDSENESGT-GHKDSVYSVVFTRDGQSVVSGS 595
Query 77 CDKTIKIWKI 86
D+++K+W +
Sbjct 596 LDRSVKLWNL 605
Score = 28.5 bits (62), Expect = 3.2, Method: Composition-based stats.
Identities = 25/99 (25%), Positives = 41/99 (41%), Gaps = 16/99 (16%)
Query 17 GDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLI-TG 75
GD+ LV GS + + WD R+G TL E+G+ A + + I G
Sbjct 497 GDK--LVSGSGDRTVRIWDLRTGQCSLTLS---------IEDGVTTVAVSPGDGKYIAAG 545
Query 76 ECDKTIKIWKIDE----EATEETHPISWGHKRDRAAVAF 110
D+ +++W + E + + GHK +V F
Sbjct 546 SLDRAVRVWDSETGFLVERLDSENESGTGHKDSVYSVVF 584
Score = 27.3 bits (59), Expect = 7.9, Method: Composition-based stats.
Identities = 16/69 (23%), Positives = 29/69 (42%), Gaps = 8/69 (11%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D L G+ + + WD + LQ E I+ + S +L++G
Sbjct 454 DGKFLATGAEDRLIRIWDIENRKIVMILQGH--------EQDIYSLDYFPSGDKLVSGSG 505
Query 78 DKTIKIWKI 86
D+T++IW +
Sbjct 506 DRTVRIWDL 514
> Hs7662316
Length=1507
Score = 36.2 bits (82), Expect = 0.016, Method: Composition-based stats.
Identities = 21/63 (33%), Positives = 31/63 (49%), Gaps = 8/63 (12%)
Query 47 SRVQPGSL-----ESENGIFCCAFDKSETRLITGECDKTIKIWKI---DEEATEETHPIS 98
SR +P S+ E E+G CCAF E L+ G C +K++ + EEA+ H +
Sbjct 1078 SRFRPISVFREANEDESGFTCCAFSARERFLMLGTCTGQLKLYNVFSGQEEASYNCHNSA 1137
Query 99 WGH 101
H
Sbjct 1138 ITH 1140
> YKR036c
Length=659
Score = 36.2 bits (82), Expect = 0.016, Method: Composition-based stats.
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 10/67 (14%)
Query 20 SSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDK 79
S+L G+ +G + WD R G + L+ +GI FD +L+TG D
Sbjct 516 SALATGTKDGIVRLWDLRVGKPVRLLEGHT--------DGITSLKFDSE--KLVTGSMDN 565
Query 80 TIKIWKI 86
+++IW +
Sbjct 566 SVRIWDL 572
Score = 28.5 bits (62), Expect = 3.1, Method: Composition-based stats.
Identities = 19/55 (34%), Positives = 25/55 (45%), Gaps = 9/55 (16%)
Query 34 WDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSE-TRLITGECDKTIKIWKID 87
WD G + L PG L + N C DK LITG D T+K+W ++
Sbjct 363 WDLNHGIQVGEL-----PGHLATVN---CMQIDKKNYNMLITGSKDATLKLWDLN 409
> CE11196
Length=439
Score = 36.2 bits (82), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 22/82 (26%), Positives = 35/82 (42%), Gaps = 6/82 (7%)
Query 6 SIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAF 65
SI + +++ D + VVG N L + G + L R E + C +
Sbjct 166 SIVKAPSKKNQLDGTEFVVGGENQLLTLYAIEKGALVEKLILRGH------ERAVECVSV 219
Query 66 DKSETRLITGECDKTIKIWKID 87
+ TR I+G D +KIW +D
Sbjct 220 NSDSTRAISGSVDTNLKIWNLD 241
> SPBC32H8.07
Length=305
Score = 35.8 bits (81), Expect = 0.018, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 41/103 (39%), Gaps = 15/103 (14%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
+CC + D+G L+ GS + FWD L+ E I F S
Sbjct 110 SCCKYVDDG---HLLTGSGDKTCMFWDIEQAKAISVLKGH--------EMDIVSLDFLPS 158
Query 69 ETRL-ITGECDKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
L +TG CDK K+W + T P G+ D +++F
Sbjct 159 NPNLFVTGGCDKLAKLWDLRAAYCCATFP---GNTSDINSISF 198
Score = 28.1 bits (61), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 18/69 (26%), Positives = 27/69 (39%), Gaps = 3/69 (4%)
Query 42 YQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTIKIWKIDEEATEETHPISWGH 101
Y T P L G C + L+TG DKT W I++ + + GH
Sbjct 90 YDTSVPDADPVELVGHAGFVSCCKYVDDGHLLTGSGDKTCMFWDIEQ---AKAISVLKGH 146
Query 102 KRDRAAVAF 110
+ D ++ F
Sbjct 147 EMDIVSLDF 155
> 7293756
Length=346
Score = 35.8 bits (81), Expect = 0.022, Method: Composition-based stats.
Identities = 27/100 (27%), Positives = 44/100 (44%), Gaps = 14/100 (14%)
Query 11 CAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSET 70
C F D+G L+ GS + ++ WD G K T+ G + +
Sbjct 155 CRFLDDG---HLITGSGDMKICHWDLEKGVK--TMDFNGHAGDIAG------LSLSPDMK 203
Query 71 RLITGECDKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
ITG DKT K+W + EE ++ + +GH D ++V +
Sbjct 204 TYITGSVDKTAKLWDVREEGHKQ---MFFGHDMDVSSVCY 240
> SPAC17H9.19c
Length=490
Score = 35.4 bits (80), Expect = 0.025, Method: Compositional matrix adjust.
Identities = 26/92 (28%), Positives = 39/92 (42%), Gaps = 11/92 (11%)
Query 4 LNSIPNCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGS------LESE 57
LN +P C F + + S L V + G L +D R Y QP + L
Sbjct 126 LNQLPFCLGFAN--NESLLAVCTETGALELFDSRF---YDRQNEENQPSARRIHGWLAHN 180
Query 58 NGIFCCAFDKSETRLITGECDKTIKIWKIDEE 89
N IF F K ++ L T D+T K++ + +
Sbjct 181 NAIFSVNFSKDDSLLATSSGDQTSKVFDLSTQ 212
> At2g47410
Length=1389
Score = 35.4 bits (80), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 12/30 (40%), Positives = 20/30 (66%), Gaps = 0/30 (0%)
Query 58 NGIFCCAFDKSETRLITGECDKTIKIWKID 87
N ++C FD+S +ITG D+ +KIW ++
Sbjct 237 NAVYCAIFDRSGRYVITGSDDRLVKIWSME 266
> Hs20357529
Length=340
Score = 35.4 bits (80), Expect = 0.028, Method: Compositional matrix adjust.
Identities = 26/102 (25%), Positives = 44/102 (43%), Gaps = 14/102 (13%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
+CC F D+ + ++ S + WD +G QT+ G + S +
Sbjct 147 SCCRFLDD---NQIITSSGDTTCALWDIETGQ--QTVGFAGHSGDVMS------LSLAPD 195
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
++G CD +IK+W + + +T GH+ D AVAF
Sbjct 196 GRTFVSGACDASIKLWDVRDSMCRQTFI---GHESDINAVAF 234
> SPAC630.14c
Length=586
Score = 35.0 bits (79), Expect = 0.037, Method: Composition-based stats.
Identities = 25/93 (26%), Positives = 38/93 (40%), Gaps = 13/93 (13%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D +LV GS + + WD +G Q L +++G+ F + G
Sbjct 387 DGKTLVSGSGDRTVCLWDVEAGE---------QKLILHTDDGVTTVMFSPDGQFIAAGSL 437
Query 78 DKTIKIWKIDEEATEETHPISWGHKRDRAAVAF 110
DK I+IW E+ H GH+ +VAF
Sbjct 438 DKVIRIWTSSGTLVEQLH----GHEESVYSVAF 466
Score = 30.8 bits (68), Expect = 0.62, Method: Composition-based stats.
Identities = 21/76 (27%), Positives = 31/76 (40%), Gaps = 10/76 (13%)
Query 18 DRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGEC 77
D L G + Q+ WD Y+ L E I+ F K L++G
Sbjct 345 DGKYLATGVEDQQIRIWDIAQKRVYRLLTGH--------EQEIYSLDFSKDGKTLVSGSG 396
Query 78 DKTIKIWKIDEEATEE 93
D+T+ +W D EA E+
Sbjct 397 DRTVCLW--DVEAGEQ 410
Score = 27.7 bits (60), Expect = 4.8, Method: Composition-based stats.
Identities = 10/30 (33%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 57 ENGIFCCAFDKSETRLITGECDKTIKIWKI 86
E ++ AF L++G D TIK+W++
Sbjct 458 EESVYSVAFSPDGKYLVSGSLDNTIKLWEL 487
> CE18673
Length=1000
Score = 34.7 bits (78), Expect = 0.039, Method: Composition-based stats.
Identities = 21/95 (22%), Positives = 43/95 (45%), Gaps = 10/95 (10%)
Query 9 NCCAFRDEGDRSSLVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKS 68
NC + G++ ++ G+++ + WD+++ QTL Q + F
Sbjct 189 NCVDYYHGGEKPYIISGADDHLVKIWDYQNKTCVQTLDGHAQ--------NVSSVCFHPE 240
Query 69 ETRLITGECDKTIKIWKIDEEATEETHPISWGHKR 103
+ITG D T+++W + E T +++G +R
Sbjct 241 LPLIITGSEDSTVRLWHANTYRLETT--LNYGLER 273
> Hs16117781
Length=707
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 20/65 (30%), Positives = 35/65 (53%), Gaps = 7/65 (10%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
LV G+ + + WD ++G QTLQ G + ++ + C F+K+ +IT D T+
Sbjct 594 LVSGNADSTVKIWDIKTGQCLQTLQ-----GPNKHQSAVTCLQFNKN--FVITSSDDGTV 646
Query 82 KIWKI 86
K+W +
Sbjct 647 KLWDL 651
> SPAC18B11.10
Length=614
Score = 34.7 bits (78), Expect = 0.040, Method: Compositional matrix adjust.
Identities = 23/89 (25%), Positives = 38/89 (42%), Gaps = 13/89 (14%)
Query 22 LVVGSNNGQLHFWDWRSGYKYQTLQSRVQPGSLESENGIFCCAFDKSETRLITGECDKTI 81
+V GS + WD +G Q ++ LE ENG+ A ++ + G D+ I
Sbjct 417 IVSGSGDRTARLWDVETG------QCILK---LEIENGVTAIAISPNDQFIAVGSLDQII 467
Query 82 KIWKIDEEATEETHPISWGHKRDRAAVAF 110
++W + E GHK ++AF
Sbjct 468 RVWSVSGTLVERLE----GHKESVYSIAF 492
Lambda K H
0.317 0.133 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1195973986
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40