bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0158_orf1
Length=317
Score E
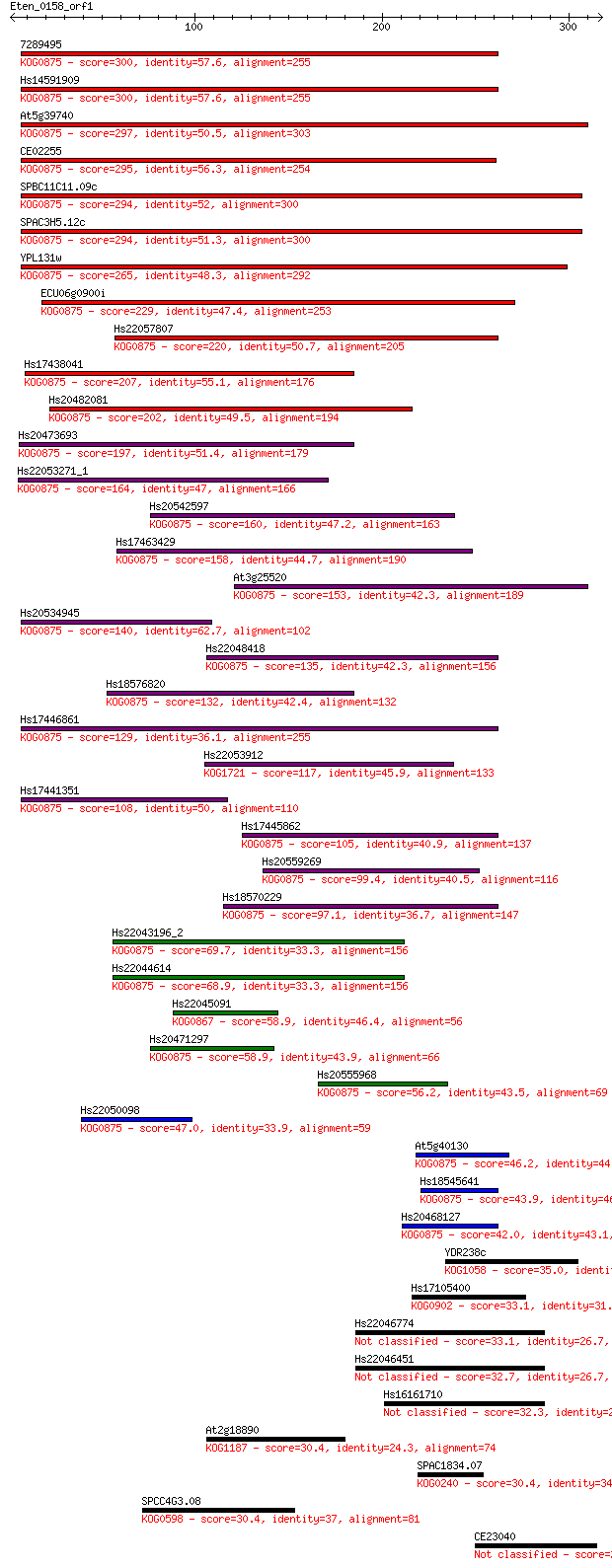
Sequences producing significant alignments: (Bits) Value
7289495 300 2e-81
Hs14591909 300 2e-81
At5g39740 297 2e-80
CE02255 295 1e-79
SPBC11C11.09c 294 1e-79
SPAC3H5.12c 294 1e-79
YPL131w 265 8e-71
ECU06g0900i 229 6e-60
Hs22057807 220 3e-57
Hs17438041 207 2e-53
Hs20482081 202 1e-51
Hs20473693 197 3e-50
Hs22053271_1 164 3e-40
Hs20542597 160 4e-39
Hs17463429 158 2e-38
At3g25520 153 4e-37
Hs20534945 140 3e-33
Hs22048418 135 1e-31
Hs18576820 132 7e-31
Hs17446861 129 1e-29
Hs22053912 117 3e-26
Hs17441351 108 2e-23
Hs17445862 105 1e-22
Hs20559269 99.4 9e-21
Hs18570229 97.1 5e-20
Hs22043196_2 69.7 8e-12
Hs22044614 68.9 1e-11
Hs22045091 58.9 1e-08
Hs20471297 58.9 1e-08
Hs20555968 56.2 8e-08
Hs22050098 47.0 5e-05
At5g40130 46.2 8e-05
Hs18545641 43.9 4e-04
Hs20468127 42.0 0.002
YDR238c 35.0 0.23
Hs17105400 33.1 0.71
Hs22046774 33.1 0.86
Hs22046451 32.7 1.1
Hs16161710 32.3 1.4
At2g18890 30.4 4.8
SPAC1834.07 30.4 4.9
SPCC4G3.08 30.4 5.0
CE23040 29.6 7.9
> 7289495
Length=319
Score = 300 bits (769), Expect = 2e-81, Method: Compositional matrix adjust.
Identities = 147/275 (53%), Positives = 191/275 (69%), Gaps = 24/275 (8%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M FVK +KNK YFKR+QVK+RRRREGKTDY AR+RL QDKNKYN PKYR +VR++N +
Sbjct 1 MGFVKVVKNKQYFKRYQVKFRRRREGKTDYYARKRLTFQDKNKYNTPKYRLIVRLSNKDI 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGI--------------------KVGLTNYSAAYA 106
Q+ YA ++GDR+VC+A S EL +YGI KVGLTNY+AAY
Sbjct 61 TVQIAYARIEGDRVVCAAYSHELPKYGIQVSFPTAFWRTQLDGPLYLFKVGLTNYAAAYC 120
Query 107 TGLLLARRLLKQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVF 166
TGLL+ARR+L + GL + G + +GEE+++E V + F+C LDVG+ TT G RVF
Sbjct 121 TGLLVARRVLNKLGLDSLYAGCTEVTGEEFNVEPVDDGPGAFRCFLDVGLARTTTGARVF 180
Query 167 GAMKGACDGGLHIPHSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDP 226
GAMKGA DGGL+IPHS KRFPG++ S+N +VHRA I+G HVA+YMR+L+EED
Sbjct 181 GAMKGAVDGGLNIPHSVKRFPGYS----AETKSFNADVHRAHIFGQHVADYMRSLEEEDE 236
Query 227 ERYQAQFSAYIRNKIDPDSIEKMYEEAFQKIRANP 261
E ++ QFS YI+ I D +E +Y++A Q IR +P
Sbjct 237 ESFKRQFSRYIKLGIRADDLEDIYKKAHQAIRNDP 271
> Hs14591909
Length=297
Score = 300 bits (769), Expect = 2e-81, Method: Compositional matrix adjust.
Identities = 147/255 (57%), Positives = 191/255 (74%), Gaps = 4/255 (1%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M FVK +KNKAYFKR+QVK+RRRREGKTDY AR+RL++QDKNKYN PKYR +VRVTN +
Sbjct 1 MGFVKVVKNKAYFKRYQVKFRRRREGKTDYYARKRLVIQDKNKYNTPKYRMIVRVTNRDI 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
+CQ+ YA ++GD +VC+A + EL +YG+KVGLTNY+AAY TGLLLARRLL + G+ ++
Sbjct 61 ICQIAYARIEGDMIVCAAYAHELPKYGVKVGLTNYAAAYCTGLLLARRLLNRFGMDKIYE 120
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G + +G+EY++E + + F C LD G+ TT GN+VFGA+KGA DGGL IPHS KRF
Sbjct 121 GQVEVTGDEYNVESIDGQPGAFTCYLDAGLARTTTGNKVFGALKGAVDGGLSIPHSTKRF 180
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
PG+ D +N EVHR I G +VA+YMR L EED + Y+ QFS YI+N + PD +
Sbjct 181 PGY----DSESKEFNAEVHRKHIMGQNVADYMRYLMEEDEDAYKKQFSQYIKNSVTPDMM 236
Query 247 EKMYEEAFQKIRANP 261
E+MY++A IR NP
Sbjct 237 EEMYKKAHAAIRENP 251
> At5g39740
Length=301
Score = 297 bits (760), Expect = 2e-80, Method: Compositional matrix adjust.
Identities = 153/305 (50%), Positives = 214/305 (70%), Gaps = 20/305 (6%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M FVK+ K+ AYFKR+QVK+RRRR+GKTDY AR RLI QDKNKYN PKYRFVVR TN +
Sbjct 1 MVFVKSSKSNAYFKRYQVKFRRRRDGKTDYRARIRLINQDKNKYNTPKYRFVVRFTNKDI 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
+ Q++ A++ GD + SA + EL +YG+ VGLTNY+AAY TGLLLARR+LK + DE++
Sbjct 61 VAQIVSASIAGDIVKASAYAHELPQYGLTVGLTNYAAAYCTGLLLARRVLKMLEMDDEYE 120
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G + +GE++ +E ++ RRPF+ +LDVG++ TT GNRVFGA+KGA DGGL IPHS+KRF
Sbjct 121 GNVEATGEDFSVEP-TDSRRPFRALLDVGLIRTTTGNRVFGALKGALDGGLDIPHSDKRF 179
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
GF K + + E+HR IYG HV+ YM+ L E++PE+ Q FSAYI+ ++ +SI
Sbjct 180 AGFHK----ENKQLDAEIHRNYIYGGHVSNYMKLLGEDEPEKLQTHFSAYIKKGVEAESI 235
Query 247 EKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVR-NV-KLDKETRKERVLK 304
E+MY++ IRA P+ K E K+A ++ R N+ KL E RK ++++
Sbjct 236 EEMYKKVHAAIRAEPNHKKTE-------------KSAPKEHKRYNLKKLTYEERKNKLIE 282
Query 305 KIQMV 309
+++ +
Sbjct 283 RVKAL 287
> CE02255
Length=293
Score = 295 bits (755), Expect = 1e-79, Method: Compositional matrix adjust.
Identities = 143/254 (56%), Positives = 183/254 (72%), Gaps = 5/254 (1%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M VK IKNKAYFKR+QVK RRRREGKTDY AR+RL +QDKNKYN PKYR +VR+TN V
Sbjct 1 MGLVKVIKNKAYFKRYQVKLRRRREGKTDYYARKRLTVQDKNKYNTPKYRLIVRITNKDV 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
+ Q+ Y+ ++GD +V SA S EL RYG+KVGLTNY+AAYATGLLLARR LK GL +K
Sbjct 61 VAQLAYSKIEGDVVVASAYSHELPRYGLKVGLTNYAAAYATGLLLARRHLKTIGLDSTYK 120
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G E+ +GE+Y++EE +R PFK VLD+G+ TT G+++F MKG DGG+++PHS RF
Sbjct 121 GHEELTGEDYNVEE-EGDRAPFKAVLDIGLARTTTGSKIFAVMKGVADGGINVPHSESRF 179
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
GF D YN E HR RI G HVA+YM LKEED +RY+ QFS ++ ++ D++
Sbjct 180 FGF----DQESKEYNAEAHRDRILGKHVADYMTYLKEEDEDRYKRQFSKFLAAGLNADNL 235
Query 247 EKMYEEAFQKIRAN 260
Y++ IRA+
Sbjct 236 VATYQKVHSAIRAD 249
> SPBC11C11.09c
Length=294
Score = 294 bits (753), Expect = 1e-79, Method: Compositional matrix adjust.
Identities = 156/300 (52%), Positives = 195/300 (65%), Gaps = 12/300 (4%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M F+KA+K+ YF R+Q KYRRRREGKTDY AR+RLI Q KNKYNAPKYR VVR +N V
Sbjct 1 MPFIKAVKSSPYFSRYQTKYRRRREGKTDYYARKRLIAQAKNKYNAPKYRLVVRFSNRFV 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
CQ++ + + GD ++ A S EL RYGIK GL N++AAYATGLL+ARR L + GLAD+++
Sbjct 61 TCQIVSSRVNGDYVLAHAHSSELPRYGIKWGLANWTAAYATGLLVARRALAKVGLADKYE 120
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G+ +P GE E + + RPFK LDVG+ T+ G+RVFGAMKGA DGGL IPHS RF
Sbjct 121 GVTEPEGEFELTEAIEDGPRPFKVFLDVGLKRTSTGSRVFGAMKGASDGGLFIPHSPNRF 180
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
PGF + DD E R IYG HVAEYM L ++D ERYQ QFS I + I+ D +
Sbjct 181 PGFDIETEELDD----ETLRKYIYGGHVAEYMEMLIDDDEERYQKQFSGLIADGIESDQL 236
Query 247 EKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVRNVKLDKETRKERVLKKI 306
E +Y EA+ KIR +P K + A K +Y + KL E RKER K+
Sbjct 237 EDIYAEAYAKIREDPSFQKSG-------KDAAAFKAESLKYTQR-KLTAEERKERFNAKV 288
> SPAC3H5.12c
Length=294
Score = 294 bits (753), Expect = 1e-79, Method: Compositional matrix adjust.
Identities = 154/300 (51%), Positives = 197/300 (65%), Gaps = 12/300 (4%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M F+KA+K+ YF R+Q KYRRRREGKTDY AR+RLI Q KNKYNAPKYR VVR +N V
Sbjct 1 MPFIKAVKSSPYFSRYQTKYRRRREGKTDYYARKRLIAQAKNKYNAPKYRLVVRFSNRFV 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
CQ++ + + GD ++ A S EL RYGIK GL N++AAYATGLL+ARR L + GLAD+++
Sbjct 61 TCQIVSSRVNGDYVLAHAHSSELPRYGIKWGLANWTAAYATGLLVARRALAKVGLADKYE 120
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G+ +P GE E + + RPFK LDVG+ T+ G+RVFGAMKGA DGGL IPHS RF
Sbjct 121 GVTEPEGEFELTEAIEDGPRPFKVFLDVGLKRTSTGSRVFGAMKGASDGGLFIPHSPNRF 180
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
PGF + DD E R IYG HVAEYM L ++D ERYQ QFS I + I+ D +
Sbjct 181 PGFDIETEELDD----ETLRKYIYGGHVAEYMEMLIDDDEERYQKQFSGLIADGIESDQL 236
Query 247 EKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVRNVKLDKETRKERVLKKI 306
E +Y EA+ KIR +P +K ++ + ++ T + KL E RKER K+
Sbjct 237 EDIYAEAYAKIREDPS-FQKSGKDAAAFKAESLKHTQR-------KLTAEERKERFNAKV 288
> YPL131w
Length=297
Score = 265 bits (678), Expect = 8e-71, Method: Compositional matrix adjust.
Identities = 141/292 (48%), Positives = 189/292 (64%), Gaps = 10/292 (3%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
MAF K K+ AY RFQ +RRRREGKTDY R+RL+ Q K KYN PKYR VVR TN +
Sbjct 1 MAFQKDAKSSAYSSRFQTPFRRRREGKTDYYQRKRLVTQHKAKYNTPKYRLVVRFTNKDI 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
+CQ++ +T+ GD ++ +A S EL RYGI GLTN++AAYATGLL+ARR L++ GL + +K
Sbjct 61 ICQIISSTITGDVVLAAAYSHELPRYGITHGLTNWAAAYATGLLIARRTLQKLGLDETYK 120
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G+E+ GE E V + RPFK LD+G+ TT G RVFGA+KGA DGGL++PHS RF
Sbjct 121 GVEEVEGEYELTEAVEDGPRPFKVFLDIGLQRTTTGARVFGALKGASDGGLYVPHSENRF 180
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
PG+ D + +PE+ R+ I+G HV++YM L ++D ER+ F Y+ + ID DS+
Sbjct 181 PGW----DFETEEIDPELLRSYIFGGHVSQYMEELADDDEERFSELFKGYLADDIDADSL 236
Query 247 EKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVRNVKLDKETR 298
E +Y A + IRA+P E + K A+S+ R KL KE R
Sbjct 237 EDIYTSAHEAIRADPAFKPTEKKFTKEQY------AAESKKYRQTKLSKEER 282
> ECU06g0900i
Length=283
Score = 229 bits (584), Expect = 6e-60, Method: Compositional matrix adjust.
Identities = 120/253 (47%), Positives = 168/253 (66%), Gaps = 6/253 (2%)
Query 18 YFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRVLCQVMYATLQG 77
Y K FQVK RRRREGKTDY R +I QD N + K+R VVR+T SRV+CQ++ A + G
Sbjct 6 YHKNFQVKRRRRREGKTDYKHRTNMIRQDSNSCGSVKHRLVVRITGSRVICQIVAAYMDG 65
Query 78 DRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFKGLEKPSGEEYH 137
DR++ ADS+EL RYGI G TNYSAAYATG L+ RR L L DE ++ G
Sbjct 66 DRVMVYADSRELERYGINFGATNYSAAYATGFLIGRRALTAMSL-DEVYRPKEIDGTYSV 124
Query 138 IEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDGAD 197
+++ E++ + LD+G+ +T G RVFGA+KGA D GL++PHS +F G DG+
Sbjct 125 TKDIDGEKKAPRVFLDIGLARSTRGARVFGALKGASDAGLNVPHSAAKFYGHK--PDGSF 182
Query 198 DSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSIEKMYEEAFQKI 257
D+ E+H RI+G +++EYM+ L++ DPE+Y+ QFS+YI+ IDP+ I ++Y+ AF KI
Sbjct 183 DA--KELH-DRIFGYNISEYMKQLRDGDPEKYKTQFSSYIKKGIDPEEIPRIYKSAFDKI 239
Query 258 RANPDPVKKEARE 270
+P KKE ++
Sbjct 240 AEDPTRKKKETKD 252
> Hs22057807
Length=247
Score = 220 bits (560), Expect = 3e-57, Method: Compositional matrix adjust.
Identities = 104/205 (50%), Positives = 143/205 (69%), Gaps = 4/205 (1%)
Query 57 FVVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLL 116
+VRVTN ++CQ+ YA ++GD +VC+A + EL +YG+KVG TNY+AAY TGL LARRLL
Sbjct 1 MIVRVTNRDIICQIAYAHIEGDMIVCTAYAHELPKYGVKVGPTNYAAAYCTGLPLARRLL 60
Query 117 KQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGG 176
+ G+ ++G + +G+EY +E + + F C LD G+ TT GN+VFGA+KGA DGG
Sbjct 61 SRFGMDKIYEGQVEVTGDEYIVESIDGQPSAFTCYLDAGLARTTTGNKVFGALKGAVDGG 120
Query 177 LHIPHSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAY 236
L IPHS K+FPG+ G + + +N EVHR I G +VA+YM L EED Y+ QFS Y
Sbjct 121 LSIPHSTKQFPGY--GSESKE--FNAEVHRKHIMGQNVADYMCYLMEEDENAYKKQFSQY 176
Query 237 IRNKIDPDSIEKMYEEAFQKIRANP 261
I+N++ PD +E+MY+EA IR NP
Sbjct 177 IKNRVTPDMMEEMYKEAHAAIRENP 201
> Hs17438041
Length=209
Score = 207 bits (528), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 97/176 (55%), Positives = 128/176 (72%), Gaps = 0/176 (0%)
Query 9 FVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRVLC 68
FVK +KNKA FKR+QVK+RRR +GKT+Y A + L++QDKNKY PKYR +VRVTN ++C
Sbjct 34 FVKVVKNKACFKRYQVKFRRRGKGKTEYHAWKHLVIQDKNKYKTPKYRMIVRVTNRDIIC 93
Query 69 QVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFKGL 128
Q+ YA + D +VC+A + EL +YG+K GLTNY AAY TGLLLA RLL + G+ ++G
Sbjct 94 QIAYAHIDRDIIVCAAYAHELPKYGMKDGLTNYVAAYCTGLLLACRLLNRFGMDKIYEGQ 153
Query 129 EKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNK 184
+ +G+EY++E + F C LD G+ TT GN+VFG +KGA DGGL IPHS K
Sbjct 154 VEVTGDEYNVESIDGPPGAFTCYLDAGLARTTTGNKVFGTLKGAVDGGLSIPHSTK 209
> Hs20482081
Length=195
Score = 202 bits (513), Expect = 1e-51, Method: Compositional matrix adjust.
Identities = 96/194 (49%), Positives = 132/194 (68%), Gaps = 4/194 (2%)
Query 22 FQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRVLCQVMYATLQGDRLV 81
+QVK+RRRREGKTDY AR+RL++QDKNKYN PKY +V V N ++CQ+ Y ++G+ +V
Sbjct 5 YQVKFRRRREGKTDYYARKRLVIQDKNKYNTPKYWMIVHVINRYIICQIAYGCIEGNVIV 64
Query 82 CSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFKGLEKPSGEEYHIEEV 141
+A + EL +YG+KVGLTNY+AAY T LLLA RLL + G+ ++G + + +EY++E +
Sbjct 65 RAAHAHELPKYGVKVGLTNYAAAYCTCLLLACRLLNRFGMDKIYEGQVEATSDEYNVESI 124
Query 142 SEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDGADDSYN 201
+ F C L G+ TT GN+VFG +K A GGL IPHS K+FP + D +N
Sbjct 125 DGQPGAFTCYLGAGLARTTTGNKVFGTLKEAAVGGLSIPHSTKQFPDY----DSESKGFN 180
Query 202 PEVHRARIYGLHVA 215
EVHR I G +VA
Sbjct 181 AEVHRKDIMGQNVA 194
> Hs20473693
Length=451
Score = 197 bits (500), Expect = 3e-50, Method: Compositional matrix adjust.
Identities = 92/179 (51%), Positives = 127/179 (70%), Gaps = 2/179 (1%)
Query 6 KMAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSR 65
++ FVK +KNKA KR+QV +RRRREGKTDY A++RL++QDKNKY+ PKYR +V VTN
Sbjct 275 RVGFVKVVKNKANLKRYQVTFRRRREGKTDYYAQKRLVMQDKNKYSTPKYRMIVCVTNRD 334
Query 66 VLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEF 125
++ ++GD +VC+A + EL +YG+KVGLTNY+AAY GLLL R+L + G+ +
Sbjct 335 II--YCLGHIEGDMIVCAAYAHELPKYGVKVGLTNYAAAYCPGLLLTHRILSRFGMDKIY 392
Query 126 KGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNK 184
+G K +G+EY +E + + F C LD G+ TT GN+VFG +KGA DGGL IP + K
Sbjct 393 EGQVKVTGDEYDVESIDGQPGAFTCYLDAGLARTTAGNQVFGTLKGAVDGGLSIPRNTK 451
> Hs22053271_1
Length=329
Score = 164 bits (414), Expect = 3e-40, Method: Compositional matrix adjust.
Identities = 78/166 (46%), Positives = 113/166 (68%), Gaps = 0/166 (0%)
Query 5 AKMAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNS 64
A + FVK +K KAY KR+QV +RR+ EGKTDY A + L++QDKNK N KYR ++ V N+
Sbjct 71 AWVGFVKVVKYKAYCKRYQVTFRRQCEGKTDYYAWKHLVVQDKNKSNTHKYRMIICVINT 130
Query 65 RVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADE 124
+C++ YA ++ D +VC+A + EL +YG+KVGLTN +AA TGLLLA RLL + G+
Sbjct 131 DTICEMAYAHIEWDMIVCAAYAHELPKYGVKVGLTNDAAACCTGLLLACRLLSRFGMDKI 190
Query 125 FKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMK 170
+KG + + +EY++ + F C LD G+ TT N+VFGA++
Sbjct 191 YKGQVEVTRDEYNVGSTDGQPGAFTCCLDAGLARTTTDNKVFGALR 236
> Hs20542597
Length=336
Score = 160 bits (404), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 77/163 (47%), Positives = 107/163 (65%), Gaps = 7/163 (4%)
Query 76 QGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFKGLEKPSGEE 135
+G+ +VC+A + EL +Y +KVGLTNY+AAY TGLLLA LL + G+ + + G + +G+E
Sbjct 178 EGEIIVCAAYAHELPKYDVKVGLTNYAAAYCTGLLLACSLLNRFGMDNIYDGQMEVTGDE 237
Query 136 YHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDG 195
Y++E + + F C LD G+ T GN+VFG +KG D L IPHS KRFPG+
Sbjct 238 YNVENIDGQPGAFTCYLDAGLARTITGNKVFGTLKGTVDRSLSIPHSTKRFPGY------ 291
Query 196 ADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIR 238
D N EVH+ RI G +VA+YMR L E+D + Y+ QFS YI+
Sbjct 292 -DSESNAEVHQKRIMGQNVADYMRYLMEQDEDAYKKQFSQYIK 333
> Hs17463429
Length=196
Score = 158 bits (399), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 85/190 (44%), Positives = 121/190 (63%), Gaps = 4/190 (2%)
Query 58 VVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLK 117
+V VT R++CQ+ Y ++GD +VC+A + +L +YG+ VGLTNY+AAY TGLLLARRLL
Sbjct 2 IVHVTKRRIICQIAYVRIEGDMIVCAAYAHKLPKYGVNVGLTNYAAAYCTGLLLARRLLN 61
Query 118 QKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGL 177
+ + ++ + +G+EY++E + + F LD + TT GN+VFGA+KG D GL
Sbjct 62 RFEVDKIYESQVEKTGDEYNVESIDGQPGAFTYYLDASLARTTTGNQVFGALKGTVDEGL 121
Query 178 HIPHSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYI 237
IPHS KRFPG+ D +N EVH+ I G VA+YM L E D + + QF Y+
Sbjct 122 SIPHSTKRFPGY----DSESKEFNAEVHQKYIMGQKVADYMHYLMEGDEDPNKKQFFQYV 177
Query 238 RNKIDPDSIE 247
+N I PD +E
Sbjct 178 KNSITPDMME 187
> At3g25520
Length=190
Score = 153 bits (387), Expect = 4e-37, Method: Compositional matrix adjust.
Identities = 80/191 (41%), Positives = 125/191 (65%), Gaps = 20/191 (10%)
Query 121 LADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIP 180
+ DE++G + +GE++ +E ++ RRPF+ +LDVG++ TT GNRVFGA+KGA DGGL IP
Sbjct 4 MDDEYEGNVEATGEDFSVEP-TDSRRPFRALLDVGLIRTTTGNRVFGALKGALDGGLDIP 62
Query 181 HSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNK 240
HS+KRF GF K + + E+HR IYG HV+ YM+ L E++PE+ Q FSAYI+
Sbjct 63 HSDKRFAGFHK----ENKQLDAEIHRNYIYGGHVSNYMKLLGEDEPEKLQTHFSAYIKKG 118
Query 241 IDPDSIEKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVR-NV-KLDKETR 298
++ +SIE++Y++ IRA+P+P K +K A Q+ R N+ KL E R
Sbjct 119 VEAESIEELYKKVHAAIRADPNPKK-------------TVKPAPKQHKRYNLKKLTYEER 165
Query 299 KERVLKKIQMV 309
K +++++++ +
Sbjct 166 KNKLIERVKAL 176
> Hs20534945
Length=123
Score = 140 bits (354), Expect = 3e-33, Method: Compositional matrix adjust.
Identities = 64/102 (62%), Positives = 82/102 (80%), Gaps = 0/102 (0%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M FVK +KNKAYFKR+QVK+RRR+EGKT Y AR+ L +QDKNKYN PKYR VV +TN +
Sbjct 1 MGFVKVVKNKAYFKRYQVKFRRRQEGKTGYYARKYLEIQDKNKYNTPKYRIVVHLTNRDI 60
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATG 108
+ Q+ YA ++GD +VC+A + EL +YG+KVGLTNY+A Y TG
Sbjct 61 ISQIAYARIEGDMIVCAAYAHELPKYGVKVGLTNYAAEYCTG 102
> Hs22048418
Length=200
Score = 135 bits (339), Expect = 1e-31, Method: Compositional matrix adjust.
Identities = 66/156 (42%), Positives = 97/156 (62%), Gaps = 7/156 (4%)
Query 106 ATGLLLARRLLKQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRV 165
+ G+LLA RLL + G+ ++G + +G EY++E + + F C LD G+ TT GN+V
Sbjct 6 SLGVLLAHRLLNRFGVDKIYEGQVEVTGNEYNVESIDGQPGAFTCYLDTGLARTTTGNKV 65
Query 166 FGAMKGACDGGLHIPHSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEED 225
FGA+KGA +G L +PHS K+FPG+ D +N EV R I +VAEY+ L E
Sbjct 66 FGALKGAVNGSLSVPHSTKQFPGY----DSESKEFNAEVQRKHIMDQNVAEYIHYLME-- 119
Query 226 PERYQAQFSAYIRNKIDPDSIEKMYEEAFQKIRANP 261
+ Y+ +FS YI+N + PD E+MY++A+ IR NP
Sbjct 120 -DAYKKEFSQYIKNSVTPDMKEEMYKKAYAAIRENP 154
> Hs18576820
Length=133
Score = 132 bits (333), Expect = 7e-31, Method: Compositional matrix adjust.
Identities = 56/132 (42%), Positives = 87/132 (65%), Gaps = 0/132 (0%)
Query 53 PKYRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLA 112
PKYR + +TN+ +C + YA ++GD ++C + EL ++G+KVGLTNY+AA+ GLLLA
Sbjct 2 PKYRMIAHITNTESVCHIAYACMEGDMIMCVPYAHELPKHGVKVGLTNYAAAHCPGLLLA 61
Query 113 RRLLKQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGA 172
RLL + G+ ++G + +G+EY++E + F C LD G+ TT GN+V G ++G+
Sbjct 62 CRLLNRSGMGKIYEGQVEVTGDEYNVESTYGQPGAFTCYLDAGLAKTTSGNKVLGGLEGS 121
Query 173 CDGGLHIPHSNK 184
DGGL PH +
Sbjct 122 TDGGLSTPHGTQ 133
> Hs17446861
Length=230
Score = 129 bits (323), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 92/255 (36%), Positives = 131/255 (51%), Gaps = 71/255 (27%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M FVK IKNKAYFKR+QVK+RRRREGKTDY A++ L D + +N +
Sbjct 1 MGFVKVIKNKAYFKRYQVKFRRRREGKTDYYAQKHL---DDSSWN-----------KQSI 46
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFK 126
+CQ+ YA +GD +VC+A EL +YG+KV LTNY+AAY TGLLL RRLL + + ++
Sbjct 47 ICQIAYARTEGDMIVCTAYVHELPKYGVKVDLTNYAAAYCTGLLLDRRLLNRFDMDKIYE 106
Query 127 GLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRF 186
G + +G++Y++ VS + +P +H H
Sbjct 107 GQVEATGDDYNV--VSIDGQP----------------------------EVHRKH----- 131
Query 187 PGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSI 246
G + AD YM L EED + Y+ QFS Y++N + PD +
Sbjct 132 ---IMGHNVAD-------------------YMCYLMEEDEDGYKKQFSQYMKNSVTPDMM 169
Query 247 EKMYEEAFQKIRANP 261
E+M ++A IR +P
Sbjct 170 EEMCKKAHAAIRESP 184
> Hs22053912
Length=687
Score = 117 bits (293), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 61/138 (44%), Positives = 83/138 (60%), Gaps = 9/138 (6%)
Query 105 YATGLLLARRLLKQK-----GLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVAT 159
Y TGLLLA RLL + G+ +G + +G EY++E + F C LDVG+ T
Sbjct 2 YCTGLLLACRLLSRSRSVTYGMDKICEGQVEVTGNEYNVESTDGQPSAFTCYLDVGLART 61
Query 160 TVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMR 219
T GN+VFG++K A DGGL IPHS K+FPG+ D +N VH+ I G +VA+YM
Sbjct 62 TTGNKVFGSLKRAVDGGLSIPHSTKQFPGY----DSESKDFNAAVHQKYIIGQNVADYMH 117
Query 220 TLKEEDPERYQAQFSAYI 237
L ++D + Y+ QFS YI
Sbjct 118 YLMKDDEDGYRKQFSQYI 135
> Hs17441351
Length=131
Score = 108 bits (269), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 55/110 (50%), Positives = 74/110 (67%), Gaps = 17/110 (15%)
Query 7 MAFVKAIKNKAYFKRFQVKYRRRREGKTDYAARRRLILQDKNKYNAPKYRFVVRVTNSRV 66
M F+K +K KAYFKR QVK+RR+ +GK DY A++RL D + VT+ +
Sbjct 1 MWFIKVVKTKAYFKRHQVKFRRQWDGKIDYYAQKRL---DDS------------VTD--I 43
Query 67 LCQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLL 116
+C + YA ++GD +VC A + EL +YG+KVGLTNY+AAY TGLLL RLL
Sbjct 44 ICHIAYACIEGDMIVCIAYAHELPKYGVKVGLTNYAAAYCTGLLLDHRLL 93
> Hs17445862
Length=146
Score = 105 bits (262), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/137 (40%), Positives = 78/137 (56%), Gaps = 9/137 (6%)
Query 125 FKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNK 184
++G + +G EY++E + + F C L + TT GN+VFGA KGA DGGL PHS+K
Sbjct 5 YEGQVEVTGNEYNVESIDGQPGAFTCYLVADLARTTTGNKVFGAPKGAVDGGLSNPHSSK 64
Query 185 RFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPD 244
RF G S +VHR I G A+ +R L EED + QFS Y +N + PD
Sbjct 65 RFLGL---------SIPHKVHRKHIMGQKFADDLRYLIEEDENASKKQFSQYTKNSVTPD 115
Query 245 SIEKMYEEAFQKIRANP 261
+E+MY++A +R NP
Sbjct 116 LMEEMYKKAHAAVRENP 132
> Hs20559269
Length=127
Score = 99.4 bits (246), Expect = 9e-21, Method: Compositional matrix adjust.
Identities = 47/116 (40%), Positives = 69/116 (59%), Gaps = 15/116 (12%)
Query 136 YHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACDGGLHIPHSNKRFPGFTKGEDG 195
Y++E + + F C LD G TT+GN+VFGA+KGA DGGL IPHS +
Sbjct 3 YNVESIDGQPGAFTCYLDAGFARTTIGNKVFGALKGAVDGGLSIPHSKE----------- 51
Query 196 ADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSIEKMYE 251
+N EVH+ I G +VA++M L EE+ + Y+ + S YI+N + D +E+M+E
Sbjct 52 ----FNEEVHQKYIIGQNVADWMHHLMEENKDAYKKELSQYIKNSVTTDVVEEMWE 103
> Hs18570229
Length=526
Score = 97.1 bits (240), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 54/148 (36%), Positives = 79/148 (53%), Gaps = 7/148 (4%)
Query 115 LLKQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACD 174
LL + G+ ++G + +G+EY++E + C LD G+ TT GN+VFG +KGA D
Sbjct 82 LLNRFGMDKIYEGQVEVTGDEYNVESTDGQPGTLTCYLDAGLARTTTGNKVFGTLKGAVD 141
Query 175 GGLHIPH-SNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQF 233
GGL +PH N K + Y RAR+ +YM L EED + Y+ Q
Sbjct 142 GGLSVPHIPNDSLVMILKARN-LMQKYTGSTSRARML-----QYMCYLMEEDEDAYKKQL 195
Query 234 SAYIRNKIDPDSIEKMYEEAFQKIRANP 261
S YI+N + D +E+MY++ IR NP
Sbjct 196 SQYIKNSVTSDMMEEMYKKGHATIRKNP 223
> Hs22043196_2
Length=415
Score = 69.7 bits (169), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 52/166 (31%), Positives = 74/166 (44%), Gaps = 52/166 (31%)
Query 56 RFVVRVTNSRVL-CQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARR 114
R +V VT +RV+ CQ+ YA +QGD N A YA
Sbjct 240 RMIVHVTPNRVIICQIAYACMQGD--------------------MNVRAPYA-------- 271
Query 115 LLKQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACD 174
+G E+ +G+EY +E + + F LD G+ TT ++VFGAM+ A D
Sbjct 272 ---------HKEGQEEVTGDEYDVESIDGQPDAFTYYLDAGLARTTTDSKVFGAME-AVD 321
Query 175 GGLHIPHSNKRFPGFTKGEDGADDSYNP---------EVHRARIYG 211
GGL IP S K+FPG+ D +N E+H +R+ G
Sbjct 322 GGLSIPQSTKQFPGY----DSESKEFNAQETVLTIHKELHNSRLGG 363
> Hs22044614
Length=455
Score = 68.9 bits (167), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 52/166 (31%), Positives = 74/166 (44%), Gaps = 52/166 (31%)
Query 56 RFVVRVTNSRVL-CQVMYATLQGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARR 114
R +V VT +RV+ CQ+ YA +QGD N A YA
Sbjct 280 RMIVHVTPNRVIICQIAYACMQGD--------------------MNVRAPYA-------- 311
Query 115 LLKQKGLADEFKGLEKPSGEEYHIEEVSEERRPFKCVLDVGIVATTVGNRVFGAMKGACD 174
+G E+ +G+EY +E + + F LD G+ TT ++VFGAM+ A D
Sbjct 312 ---------HKEGQEEVTGDEYDVESIDGQPDAFTYYLDAGLARTTTDSKVFGAME-AVD 361
Query 175 GGLHIPHSNKRFPGFTKGEDGADDSYNP---------EVHRARIYG 211
GGL IP S K+FPG+ D +N E+H +R+ G
Sbjct 362 GGLSIPQSTKQFPGY----DSESKEFNAQETVLTIHKELHNSRLGG 403
> Hs22045091
Length=319
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 26/56 (46%), Positives = 41/56 (73%), Gaps = 0/56 (0%)
Query 88 ELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFKGLEKPSGEEYHIEEVSE 143
EL +YG+KVGLT+Y+ AY TGLLLA RLL + G+ ++G + + +E+++E + E
Sbjct 223 ELPKYGVKVGLTHYATAYCTGLLLACRLLSKSGMVKIYEGQVEVTRDEHNVENIDE 278
> Hs20471297
Length=395
Score = 58.9 bits (141), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 29/66 (43%), Positives = 44/66 (66%), Gaps = 0/66 (0%)
Query 76 QGDRLVCSADSQELTRYGIKVGLTNYSAAYATGLLLARRLLKQKGLADEFKGLEKPSGEE 135
+GD +VC+A + EL +YG+K GLTN +A Y T LLLA RLL + G+ + +G+E
Sbjct 253 EGDMIVCTAYAHELPKYGMKFGLTNDAAMYCTVLLLAHRLLNKFGMEKICEHQVAVTGDE 312
Query 136 YHIEEV 141
Y++E +
Sbjct 313 YNVEGI 318
> Hs20555968
Length=91
Score = 56.2 bits (134), Expect = 8e-08, Method: Composition-based stats.
Identities = 30/69 (43%), Positives = 39/69 (56%), Gaps = 4/69 (5%)
Query 166 FGAMKGACDGGLHIPHSNKRFPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEED 225
+G KG DGGL IPH+ KRFPG+ D +N EVH+ I + A YM L E+D
Sbjct 27 YGVWKGVVDGGLLIPHNIKRFPGY----DSESKKFNAEVHQKPIMDPNAAGYMCYLIEKD 82
Query 226 PERYQAQFS 234
+ Y+ FS
Sbjct 83 EDAYKKCFS 91
> Hs22050098
Length=247
Score = 47.0 bits (110), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 20/59 (33%), Positives = 35/59 (59%), Gaps = 0/59 (0%)
Query 39 RRRLILQDKNKYNAPKYRFVVRVTNSRVLCQVMYATLQGDRLVCSADSQELTRYGIKVG 97
+ L L D+ + A +V +TN V C ++YA ++GD +VC+A + +L RYG+ +
Sbjct 133 HQNLPLNDETELEASHLMRIVDITNRDVNCYLVYAGIEGDMIVCTAHAHKLPRYGVNLA 191
> At5g40130
Length=88
Score = 46.2 bits (108), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 22/50 (44%), Positives = 33/50 (66%), Gaps = 0/50 (0%)
Query 218 MRTLKEEDPERYQAQFSAYIRNKIDPDSIEKMYEEAFQKIRANPDPVKKE 267
M+ L E +PE+ Q FSAYI+ ++ +SIE+MY++ IRA P+ K E
Sbjct 1 MKLLGEYEPEKLQTLFSAYIKKGVEAESIEEMYKKVHAAIRAEPNHKKTE 50
> Hs18545641
Length=105
Score = 43.9 bits (102), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/41 (46%), Positives = 28/41 (68%), Gaps = 0/41 (0%)
Query 221 LKEEDPERYQAQFSAYIRNKIDPDSIEKMYEEAFQKIRANP 261
L EED + Y+ QFS YI+N + PD +E+MY++A + NP
Sbjct 26 LLEEDEDAYKKQFSQYIKNSVTPDMMEEMYKKAHAAVPENP 66
> Hs20468127
Length=91
Score = 42.0 bits (97), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/51 (43%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Query 211 GLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDSIEKMYEEAFQKIRANP 261
G +VA+YM L +ED + Y+ QFS YI+N + PD +K A Q+ NP
Sbjct 2 GQNVADYMCYLMQEDEDAYKKQFSQYIKNSVTPDMRDKKAHAALQE---NP 49
> YDR238c
Length=973
Score = 35.0 bits (79), Expect = 0.23, Method: Composition-based stats.
Identities = 21/72 (29%), Positives = 39/72 (54%), Gaps = 5/72 (6%)
Query 234 SAYIRNKIDPDSIEK-MYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVRNVK 292
S+ + KID DS+E+ M + NP+ K+E ++ + A + T KS + R ++
Sbjct 608 SSLVEKKIDEDSLERVMTSISILLDEVNPEEKKEEV----KLLEVAFLDTTKSSFKRQIE 663
Query 293 LDKETRKERVLK 304
+ K+ + +R LK
Sbjct 664 IAKKNKHKRALK 675
> Hs17105400
Length=2044
Score = 33.1 bits (74), Expect = 0.71, Method: Composition-based stats.
Identities = 19/70 (27%), Positives = 31/70 (44%), Gaps = 9/70 (12%)
Query 216 EYMRTLKEEDPERYQAQFSAYIRNKIDPD---------SIEKMYEEAFQKIRANPDPVKK 266
EYMR L+ DP+R+Q F + I D ++ +AF + A+ K+
Sbjct 842 EYMRVLRSTDPDRFQVMFCYFEDKAIQKDKSGMMQCVIAVADKVFDAFLNMMADKAKTKE 901
Query 267 EAREVKRVRQ 276
E++R Q
Sbjct 902 NEEELERHAQ 911
> Hs22046774
Length=994
Score = 33.1 bits (74), Expect = 0.86, Method: Composition-based stats.
Identities = 27/108 (25%), Positives = 47/108 (43%), Gaps = 11/108 (10%)
Query 186 FPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDS 245
FP K E N E H R+ GL + K ED +Q + + +K P S
Sbjct 751 FPPIHKSEKSRKP--NLEKHEERLEGLRTPQLTPVRKTEDT--HQDEGVQLLPSKKQPPS 806
Query 246 -------IEKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQ 286
I+++++ F K ++ P PV E+++ + R +A++Q
Sbjct 807 VSPFGENIKQIFQWIFSKKKSKPAPVTAESQKTVKNRSRVYSSSAEAQ 854
> Hs22046451
Length=990
Score = 32.7 bits (73), Expect = 1.1, Method: Composition-based stats.
Identities = 27/108 (25%), Positives = 46/108 (42%), Gaps = 11/108 (10%)
Query 186 FPGFTKGEDGADDSYNPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDS 245
FP K E N E H R+ GL + K ED +Q + + +K P S
Sbjct 747 FPPIHKSEKSRKP--NLEKHEERLEGLRTPQLTPVRKTEDT--HQDEGVQLLPSKKQPPS 802
Query 246 -------IEKMYEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQ 286
I++ ++ F K ++ P PV E+++ + R +A++Q
Sbjct 803 VSHFGENIKQFFQWIFSKKKSKPAPVTAESQKTVKNRSCVYSSSAEAQ 850
> Hs16161710
Length=994
Score = 32.3 bits (72), Expect = 1.4, Method: Composition-based stats.
Identities = 23/93 (24%), Positives = 43/93 (46%), Gaps = 9/93 (9%)
Query 201 NPEVHRARIYGLHVAEYMRTLKEEDPERYQAQFSAYIRNKIDPDS-------IEKMYEEA 253
N E H R+ GL + K ED +Q + + +K P S I+++++
Sbjct 764 NLEKHEERLEGLRTPQLTPVRKTEDT--HQDEGVQLLPSKKQPPSVSPFGENIKQIFQWI 821
Query 254 FQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQ 286
F K ++ P PV E+++ + R +A++Q
Sbjct 822 FSKKKSKPAPVTAESQKTVKNRSCVYSSSAEAQ 854
> At2g18890
Length=392
Score = 30.4 bits (67), Expect = 4.8, Method: Compositional matrix adjust.
Identities = 18/79 (22%), Positives = 39/79 (49%), Gaps = 5/79 (6%)
Query 106 ATGLLLARRLLKQKGLADEFKGLEKPSGEEYHIEEVS-----EERRPFKCVLDVGIVATT 160
AT + L+ + G A+ +KG+ +GEE ++ ++ +ERR + ++++G +
Sbjct 64 ATNGFSSENLVGRGGFAEVYKGILGKNGEEIAVKRITRGGRDDERREKEFLMEIGTIGHV 123
Query 161 VGNRVFGAMKGACDGGLHI 179
V + D GL++
Sbjct 124 SHPNVLSLLGCCIDNGLYL 142
> SPAC1834.07
Length=554
Score = 30.4 bits (67), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 12/35 (34%), Positives = 23/35 (65%), Gaps = 0/35 (0%)
Query 219 RTLKEEDPERYQAQFSAYIRNKIDPDSIEKMYEEA 253
RT++E + +R ++ ++ R+ D DSI ++Y EA
Sbjct 401 RTIQESNNDRDESTVASIHRHNFDSDSINRLYAEA 435
> SPCC4G3.08
Length=436
Score = 30.4 bits (67), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 30/87 (34%), Positives = 39/87 (44%), Gaps = 9/87 (10%)
Query 72 YATLQGDRLVCS-----ADSQELTRYG-IKVGLTNYSAAYATGLLLARRLLKQKGLADEF 125
+ + +GD V S AD Q LT G G TG L A++ LK+ +
Sbjct 72 FISSKGDNYVPSGKMRPADFQPLTVLGRGSYGKVLLVKQKNTGRLFAQKQLKKASIVLRA 131
Query 126 KGLEKPSGEEYHIEEVSEERRPFKCVL 152
KGLE+ E +EEV R PF C L
Sbjct 132 KGLEQTKNERQILEEV---RHPFICRL 155
> CE23040
Length=340
Score = 29.6 bits (65), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 23/68 (33%), Positives = 38/68 (55%), Gaps = 5/68 (7%)
Query 250 YEEAFQKIRANPDPVKKEAREVKRVRQGAMIKTAKSQYVRNVKLDK---ETRKERVLKKI 306
+EE F+K+ N DP K + RE +R RQ A + + V+ K + E R ++ K+I
Sbjct 97 WEEEFKKL--NKDPSKMDKREQERHRQMAKNREYARKCVQKKKDQRKHAEIRSNQIKKRI 154
Query 307 QMVADKMA 314
Q++ +K A
Sbjct 155 QLLKNKTA 162
Lambda K H
0.319 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7378771334
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40