bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0155_orf3
Length=372
Score E
Sequences producing significant alignments: (Bits) Value
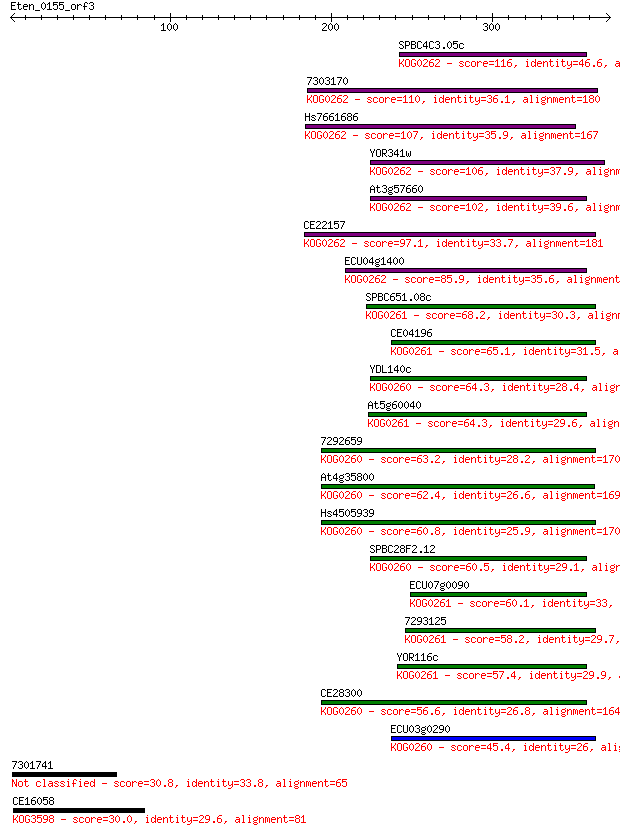
SPBC4C3.05c 116 1e-25
7303170 110 6e-24
Hs7661686 107 4e-23
YOR341w 106 8e-23
At3g57660 102 2e-21
CE22157 97.1 6e-20
ECU04g1400 85.9 1e-16
SPBC651.08c 68.2 3e-11
CE04196 65.1 2e-10
YDL140c 64.3 4e-10
At5g60040 64.3 4e-10
7292659 63.2 9e-10
At4g35800 62.4 1e-09
Hs4505939 60.8 5e-09
SPBC28F2.12 60.5 6e-09
ECU07g0090 60.1 7e-09
7293125 58.2 3e-08
YOR116c 57.4 5e-08
CE28300 56.6 8e-08
ECU03g0290 45.4 2e-04
7301741 30.8 5.4
CE16058 30.0 8.8
> SPBC4C3.05c
Length=1689
Score = 116 bits (290), Expect = 1e-25, Method: Compositional matrix adjust.
Identities = 54/116 (46%), Positives = 75/116 (64%), Gaps = 0/116 (0%)
Query 242 VDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAI 301
+ + + TND+ A+L+ YG+EAAR + E S VF YGI V HL LIAD++T +G
Sbjct 1559 ISMNDIYTNDIAAILRIYGVEAARNAIVHEVSSVFGVYGIAVDPRHLSLIADYMTFEGGY 1618
Query 302 IPFNRLGIQHCDSPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGS 357
FNR+GI++ SPF +M+FET+ FLTEA R VD+L PS+ L+VG G+
Sbjct 1619 KAFNRMGIEYNTSPFAKMSFETTCHFLTEAALRGDVDDLSNPSSRLVVGRVGNFGT 1674
> 7303170
Length=1642
Score = 110 bits (274), Expect = 6e-24, Method: Compositional matrix adjust.
Identities = 65/180 (36%), Positives = 97/180 (53%), Gaps = 5/180 (2%)
Query 185 ELLPIVKGLVEKVTLQEPQGVSNPRIVQGSGCSTGSPMELHCSGSNLQWLHRLKETAVDH 244
+L I++ L K + + Q + I +G T L G N+ + + +D
Sbjct 1465 DLTSIIRELAGKSVVHQVQHIKRAIIYKG----TDDDQLLKTDGINIGEMFQ-HNKILDL 1519
Query 245 SKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPF 304
++L +ND+ A+ + YGIEAA + +KE S VF YGI V HL LIAD++T DG P
Sbjct 1520 NRLYSNDIHAIARTYGIEAASQVIVKEVSNVFKVYGITVDRRHLSLIADYMTFDGTFQPL 1579
Query 305 NRLGIQHCDSPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGSHQSQLLA 364
+R G++H SP QM+FE+S +FL A D L +PS+ L+VG P G+ +LL
Sbjct 1580 SRKGMEHSSSPLQQMSFESSLQFLKSAAGFGRADELSSPSSRLMVGLPVRNGTGAFELLT 1639
> Hs7661686
Length=1717
Score = 107 bits (267), Expect = 4e-23, Method: Compositional matrix adjust.
Identities = 60/167 (35%), Positives = 90/167 (53%), Gaps = 2/167 (1%)
Query 184 LELLPIVKGLVEKVTLQEPQGVSNPRIVQGSGCSTGSPMELHCSGSNLQWLHRLKETAVD 243
++ +V L + +G++ + + + + L+ G NL L + E +D
Sbjct 1536 FDMSSLVVSLAHGAVIYATKGITRCLLNETTNNKNEKELVLNTEGINLPELFKYAEV-LD 1594
Query 244 HSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIP 303
+L +ND+ A+ YGIEA R + KE VFA YGI V HL L+AD++ +G P
Sbjct 1595 LRRLYSNDIHAIANTYGIEALRVIE-KEIKDVFAVYGIAVDPRHLSLVADYMCFEGVYKP 1653
Query 304 FNRLGIQHCDSPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVG 350
NR GI+ SP QM FETSF+FL +A + D L++PSA L+VG
Sbjct 1654 LNRFGIRSNSSPLQQMTFETSFQFLKQATMLGSHDELRSPSACLVVG 1700
> YOR341w
Length=1664
Score = 106 bits (265), Expect = 8e-23, Method: Compositional matrix adjust.
Identities = 55/145 (37%), Positives = 87/145 (60%), Gaps = 1/145 (0%)
Query 224 LHCSGSNLQWLHRLKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKV 283
L G N Q + +E +D + +NDV A+L+ YG+EAAR + E + VF+ Y I V
Sbjct 1519 LVTEGVNFQAMWD-QEAFIDVDGITSNDVAAVLKTYGVEAARNTIVNEINNVFSRYAISV 1577
Query 284 HSSHLGLIADFVTRDGAIIPFNRLGIQHCDSPFLQMAFETSFRFLTEACERSAVDNLKTP 343
HL LIAD +TR G + FNR G++ S F++M++ET+ +FLT+A + + L +P
Sbjct 1578 SFRHLDLIADMMTRQGTYLAFNRQGMETSTSSFMKMSYETTCQFLTKAVLDNEREQLDSP 1637
Query 344 SAALIVGSPAVIGSHQSQLLAVLPD 368
SA ++VG +G+ +LA +P+
Sbjct 1638 SARIVVGKLNNVGTGSFDVLAKVPN 1662
> At3g57660
Length=1670
Score = 102 bits (253), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 53/135 (39%), Positives = 79/135 (58%), Gaps = 2/135 (1%)
Query 224 LHCSGSNLQWLHRLKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKV 283
LH SG + L ++ +D L +N + ML +G+EAAR ++E + VF +YGI V
Sbjct 1526 LHASGVDFPALWEFQD-KLDVRYLYSNSIHDMLNIFGVEAARETIIREINHVFKSYGISV 1584
Query 284 HSSHLGLIADFVTRDGAIIPFNRL-GIQHCDSPFLQMAFETSFRFLTEACERSAVDNLKT 342
HL LIAD++T G P +R+ GI SPF +M FET+ +F+ +A D L+T
Sbjct 1585 SIRHLNLIADYMTFSGGYRPMSRMGGIAESTSPFCRMTFETATKFIVQAATYGEKDTLET 1644
Query 343 PSAALIVGSPAVIGS 357
PSA + +G PA+ G+
Sbjct 1645 PSARICLGLPALSGT 1659
> CE22157
Length=1737
Score = 97.1 bits (240), Expect = 6e-20, Method: Compositional matrix adjust.
Identities = 61/182 (33%), Positives = 95/182 (52%), Gaps = 4/182 (2%)
Query 183 RLELLPIVKGLVEKVTLQEPQGVSNPRIVQGSGCSTGSPME-LHCSGSNLQWLHRLKETA 241
++++ IV+ VE + + G+ R V+ + G M L G NL + +
Sbjct 1533 KMDVSSIVEKEVELFIVHQTPGIE--RCVETTEQKNGKEMTILQTQGVNLAAFFKHADV- 1589
Query 242 VDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAI 301
+D + + +ND+ +L++YG+EA E + VFA YGI+V HL L AD++T G I
Sbjct 1590 LDVNSVYSNDLNLILENYGVEACSKAITTEMNNVFAVYGIEVSKRHLSLTADYMTFTGQI 1649
Query 302 IPFNRLGIQHCDSPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGSHQSQ 361
PFNR + SP +M FET+ FL EA DN+ +PSA L++G+ G+
Sbjct 1650 QPFNRGAMSSSSSPLQKMTFETTMAFLREALLHGEEDNVNSPSARLVMGALPRGGTGSFD 1709
Query 362 LL 363
LL
Sbjct 1710 LL 1711
> ECU04g1400
Length=1395
Score = 85.9 bits (211), Expect = 1e-16, Method: Composition-based stats.
Identities = 53/159 (33%), Positives = 82/159 (51%), Gaps = 10/159 (6%)
Query 209 RIVQGSGCSTGSPMELHCSGSNLQWLHRLKETA----------VDHSKLETNDVLAMLQH 258
R V+G ++ S +L S++ L R+ E + +D E+ND+ +
Sbjct 1222 REVKGMEKASVSGSQLFVKSSSIYSLTRMIEVSPGVYEDLLDILDIYNAESNDIYDVYLT 1281
Query 259 YGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFNRLGIQHCDSPFLQ 318
G+EAAR + E +VF YGI + HL LIAD++TR G+ PF+R G+ DSP +
Sbjct 1282 LGVEAARHAIINEVVKVFDVYGISIDIRHLLLIADYMTRKGSYSPFSRHGLGADDSPIQR 1341
Query 319 MAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGS 357
++FE+ + A D L PSA+L VG+P G+
Sbjct 1342 ISFESCYSNFKTAATFHLEDKLSNPSASLTVGNPVRCGT 1380
> SPBC651.08c
Length=1405
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 43/143 (30%), Positives = 70/143 (48%), Gaps = 3/143 (2%)
Query 222 MELHCSGSNLQWLHRLKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGI 281
+EL G+ LQ + + + +K TN V+ M GIEAAR + E A +G+
Sbjct 1237 IELFMEGTGLQAV--MNTEGIVGTKTSTNHVMEMKDVLGIEAARYSIISEIGYTMAKHGL 1294
Query 282 KVHSSHLGLIADFVTRDGAIIPFNRLGIQHCDSPFLQMA-FETSFRFLTEACERSAVDNL 340
V H+ L+ D +T G ++ R G+ L +A FE + L A R A D++
Sbjct 1295 TVDPRHIMLLGDVMTCKGEVLGITRFGVAKMKDSVLALASFEKTTDHLFNAAARFAKDSI 1354
Query 341 KTPSAALIVGSPAVIGSHQSQLL 363
+ S +++G A IG++ QL+
Sbjct 1355 EGISECIVLGKLAPIGTNVFQLI 1377
> CE04196
Length=1388
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 40/128 (31%), Positives = 63/128 (49%), Gaps = 1/128 (0%)
Query 237 LKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVT 296
L VD K N+ L + GIEAAR+ + E A+GI + H+ L+AD +T
Sbjct 1238 LSSVGVDPRKTNFNNALVVADVLGIEAARSCIINEIIATMDAHGIGLDRRHVMLLADVMT 1297
Query 297 RDGAIIPFNRLG-IQHCDSPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVI 355
G ++ R G ++ DS L +FE + L EA S D + S +I+G+P +
Sbjct 1298 YRGEVLGITRNGLVKMKDSVLLLASFEKTMDHLFEAAFFSQRDVIHGVSECIIMGTPMTV 1357
Query 356 GSHQSQLL 363
G+ +L+
Sbjct 1358 GTGTFKLM 1365
> YDL140c
Length=1733
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 38/135 (28%), Positives = 67/135 (49%), Gaps = 3/135 (2%)
Query 224 LHCSGSNLQWLHRLKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKV 283
L G NL + + +D +++ TN + +++ GIEA RA KE V A+ G V
Sbjct 1306 LETDGVNLSEV--MTVPGIDPTRIYTNSFIDIMEVLGIEAGRAALYKEVYNVIASDGSYV 1363
Query 284 HSSHLGLIADFVTRDGAIIPFNRLGIQHCDS-PFLQMAFETSFRFLTEACERSAVDNLKT 342
+ H+ L+ D +T G + R G ++ ++ +FE + L EA + +D+ +
Sbjct 1364 NYRHMALLVDVMTTQGGLTSVTRHGFNRSNTGALMRCSFEETVEILFEAGASAELDDCRG 1423
Query 343 PSAALIVGSPAVIGS 357
S +I+G A IG+
Sbjct 1424 VSENVILGQMAPIGT 1438
> At5g60040
Length=1328
Score = 64.3 bits (155), Expect = 4e-10, Method: Composition-based stats.
Identities = 40/136 (29%), Positives = 69/136 (50%), Gaps = 3/136 (2%)
Query 223 ELHCSGSNLQWLHRLKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIK 282
+L G+NL L + ++ +N+V+ + + GIEAAR + E V +G+
Sbjct 1171 KLFVEGTNL--LAVMGTPGINGRTTTSNNVVEVSKTLGIEAARTTIIDEIGTVMGNHGMS 1228
Query 283 VHSSHLGLIADFVTRDGAIIPFNRLGIQHCD-SPFLQMAFETSFRFLTEACERSAVDNLK 341
+ H+ L+AD +T G ++ R GIQ D S +Q +FE + L A VDN++
Sbjct 1229 IDIRHMMLLADVMTYRGEVLGIQRTGIQKMDKSVLMQASFERTGDHLFSAAASGKVDNIE 1288
Query 342 TPSAALIVGSPAVIGS 357
+ +I+G P +G+
Sbjct 1289 GVTECVIMGIPMKLGT 1304
> 7292659
Length=1887
Score = 63.2 bits (152), Expect = 9e-10, Method: Composition-based stats.
Identities = 48/171 (28%), Positives = 76/171 (44%), Gaps = 1/171 (0%)
Query 194 VEKVTLQEPQGVSNPRIVQGSGCSTGSPMELHCSGSNLQWLHRLKETAVDHSKLETNDVL 253
+ KV + PQ S RIV + E + L E VD + +ND+
Sbjct 1296 IGKVYMHLPQTDSKKRIVITETGEFKAIGEWLLETDGTSMMKVLSERDVDPIRTSSNDIC 1355
Query 254 AMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFNRLGIQHCD 313
+ Q GIEA R KE + V YG+ V+ HL L+ D +T G ++ R GI D
Sbjct 1356 EIFQVLGIEAVRKSVEKEMNAVLQFYGLYVNYRHLALLCDVMTAKGHLMAITRHGINRQD 1415
Query 314 S-PFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGSHQSQLL 363
+ ++ +FE + L +A + D ++ S +I+G +G+ LL
Sbjct 1416 TGALMRCSFEETVDVLMDAAAHAETDPMRGVSENIIMGQLPKMGTGCFDLL 1466
> At4g35800
Length=1840
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 45/170 (26%), Positives = 79/170 (46%), Gaps = 6/170 (3%)
Query 194 VEKVTLQEPQGVSNPRIVQGSGCSTGSPMELHCSGSNLQWLHRLKETAVDHSKLETNDVL 253
+ KV +++ V R + G T L G NL L + VD + +N ++
Sbjct 1301 INKVFIKQ---VRKSRFDEEGGFKTSEEWMLDTEGVNL--LAVMCHEDVDPKRTTSNHLI 1355
Query 254 AMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFNRLGIQHCD 313
+++ GIEA R L E V + G V+ HL ++ D +T G ++ R GI D
Sbjct 1356 EIIEVLGIEAVRRALLDELRVVISFDGSYVNYRHLAILCDTMTYRGHLMAITRHGINRND 1415
Query 314 S-PFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGSHQSQL 362
+ P ++ +FE + L +A + D L+ + +++G A IG+ +L
Sbjct 1416 TGPLMRCSFEETVDILLDAAAYAETDCLRGVTENIMLGQLAPIGTGDCEL 1465
> Hs4505939
Length=1970
Score = 60.8 bits (146), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 44/171 (25%), Positives = 75/171 (43%), Gaps = 1/171 (0%)
Query 194 VEKVTLQEPQGVSNPRIVQGSGCSTGSPMELHCSGSNLQWLHRLKETAVDHSKLETNDVL 253
+ KV + PQ + +I+ + E + + L E VD + +ND++
Sbjct 1304 ISKVYMHLPQTDNKKKIIITEDGEFKALQEWILETDGVSLMRVLSEKDVDPVRTTSNDIV 1363
Query 254 AMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFNRLGIQHCD 313
+ GIEA R +E V + G V+ HL L+ D +T G ++ R G+ D
Sbjct 1364 EIFTVLGIEAVRKALERELYHVISFDGSYVNYRHLALLCDTMTCRGHLMAITRHGVNRQD 1423
Query 314 S-PFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGSHQSQLL 363
+ P ++ +FE + L EA D +K S +++G A G+ LL
Sbjct 1424 TGPLMKCSFEETVDVLMEAAAHGESDPMKGVSENIMLGQLAPAGTGCFDLL 1474
> SPBC28F2.12
Length=1752
Score = 60.5 bits (145), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 64/135 (47%), Gaps = 3/135 (2%)
Query 224 LHCSGSNLQWLHRLKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKV 283
L G NL + VD ++ +N + +LQ GIEA R+ LKE V G V
Sbjct 1312 LETDGINLT--EAMTVEGVDATRTYSNSFVEILQILGIEATRSALLKELRNVIEFDGSYV 1369
Query 284 HSSHLGLIADFVTRDGAIIPFNRLGIQHCDS-PFLQMAFETSFRFLTEACERSAVDNLKT 342
+ HL L+ D +T G ++ R GI ++ ++ +FE + L +A D+ K
Sbjct 1370 NYRHLALLCDVMTSRGHLMAITRHGINRAETGALMRCSFEETVEILMDAAASGEKDDCKG 1429
Query 343 PSAALIVGSPAVIGS 357
S +++G A +G+
Sbjct 1430 ISENIMLGQLAPMGT 1444
> ECU07g0090
Length=1349
Score = 60.1 bits (144), Expect = 7e-09, Method: Composition-based stats.
Identities = 36/110 (32%), Positives = 57/110 (51%), Gaps = 1/110 (0%)
Query 249 TNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFNRLG 308
+N + + + GIEAARA + E + +GIK+ H+ L+AD +T G + R G
Sbjct 1229 SNSITEVEEVLGIEAARAQIIHEIEYTISNHGIKIDPRHIMLLADTMTYRGEVFGITRFG 1288
Query 309 IQHCDSPFLQMA-FETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGS 357
I L +A FE + +L EA +S D + S ++I+G P IG+
Sbjct 1289 ISKMSRSTLMLASFEQTSDYLFEAAVQSKSDEICGVSESIILGIPICIGT 1338
> 7293125
Length=1259
Score = 58.2 bits (139), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 35/119 (29%), Positives = 60/119 (50%), Gaps = 1/119 (0%)
Query 246 KLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFN 305
+ +N++ + Q GIEAAR + + E + V +G+ V H+ L+A +T G ++
Sbjct 1114 RTRSNNICEIYQTLGIEAARTIIMSEITEVMEGHGMSVDWRHIMLLASQMTARGEVLGIT 1173
Query 306 RLGIQHC-DSPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGSHQSQLL 363
R G+ +S F +FE + L +A D + S +I+G PA IG+ +LL
Sbjct 1174 RHGLAKMRESVFNLASFEKTADHLFDAAYYGQTDAINGVSERIILGMPACIGTGIFKLL 1232
> YOR116c
Length=1460
Score = 57.4 bits (137), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 35/118 (29%), Positives = 57/118 (48%), Gaps = 1/118 (0%)
Query 241 AVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGA 300
V S+ TN VL + GIEAAR ++E + + +G+ V H+ L+ D +T G
Sbjct 1308 GVIGSRTTTNHVLEVFSVLGIEAARYSIIREINYTMSNHGMSVDPRHIQLLGDVMTYKGE 1367
Query 301 IIPFNRLGIQHCDSPFLQMA-FETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGS 357
++ R G+ LQ+A FE + L +A D ++ S +I+G IG+
Sbjct 1368 VLGITRFGLSKMRDSVLQLASFEKTTDHLFDAAFYMKKDAVEGVSECIILGQTMSIGT 1425
> CE28300
Length=1852
Score = 56.6 bits (135), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 44/167 (26%), Positives = 75/167 (44%), Gaps = 5/167 (2%)
Query 194 VEKVTLQEPQGVSNPRIVQG--SGCSTGSPMELHCSGSNLQWLHRLKETAVDHSKLETND 251
+ KV + +P RI+ G + + L G+ L L L E +D + +ND
Sbjct 1292 ISKVYMNQPNTDDKKRIIITPEGGFKSVADWILETDGTAL--LRVLSERQIDPVRTTSND 1349
Query 252 VLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVTRDGAIIPFNRLGIQH 311
+ + + GIEA R +E V + G V+ HL L+ D +T G ++ R GI
Sbjct 1350 ICEIFEVLGIEAVRKAIEREMDNVISFDGSYVNYRHLALLCDVMTAKGHLMAITRHGINR 1409
Query 312 CD-SPFLQMAFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVIGS 357
+ ++ +FE + L EA + D +K S +++G A G+
Sbjct 1410 QEVGALMRCSFEETVDILMEAAVHAEEDPVKGVSENIMLGQLARCGT 1456
> ECU03g0290
Length=1599
Score = 45.4 bits (106), Expect = 2e-04, Method: Composition-based stats.
Identities = 33/128 (25%), Positives = 63/128 (49%), Gaps = 1/128 (0%)
Query 237 LKETAVDHSKLETNDVLAMLQHYGIEAARALYLKEFSRVFAAYGIKVHSSHLGLIADFVT 296
L AV+ +ND++ + + GIEAAR L+E + V G V+ H+ L+AD +T
Sbjct 1280 LGNPAVNSRLTISNDLVEIAETLGIEAARESILRELTIVIDGNGSYVNYRHMSLLADVMT 1339
Query 297 RDGAIIPFNRLGIQHCDSPFLQM-AFETSFRFLTEACERSAVDNLKTPSAALIVGSPAVI 355
G + R G+ + L+ +FE + L +A S + + + +++G A +
Sbjct 1340 MRGYLCGITRHGVNKVGAGALKRSSFEETVEILLDAALVSEKNICRGITENIMMGQLAPM 1399
Query 356 GSHQSQLL 363
G+ +++
Sbjct 1400 GTGNIEIM 1407
> 7301741
Length=2977
Score = 30.8 bits (68), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 13/65 (20%)
Query 2 TREDEEQQQQQQQQQQQQQESESDSSSEEEEANVKQEKGEAAKVKREIKEEEFEANKQPI 61
T E E +QQ +++Q ++ +ESD SE AKVK EIK+E + + QP
Sbjct 2273 TAEKELEQQLKEEQTSEKSTAESDVLSE-------------AKVKEEIKQEPIDKSAQPD 2319
Query 62 KKIKK 66
+ K
Sbjct 2320 GVVDK 2324
> CE16058
Length=3498
Score = 30.0 bits (66), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 24/86 (27%), Positives = 52/86 (60%), Gaps = 5/86 (5%)
Query 3 REDEEQQQQQQQQ-----QQQQQESESDSSSEEEEANVKQEKGEAAKVKREIKEEEFEAN 57
RE+EE+ +Q+Q + +++ + +E + +E E V++EK EAA+ ++E +E+E
Sbjct 2649 RENEERVRQEQMRLEAEERERIRRAEEERIQKELEDKVRREKEEAARQEKERQEQEARMR 2708
Query 58 KQPIKKIKKEQQQQQQQQQQQPQQQQ 83
+ ++ +++ +QQ++ QQ P Q
Sbjct 2709 EAREAELSRQRMEQQRRSQQNPYMNQ 2734
Lambda K H
0.314 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9190772932
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40