bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
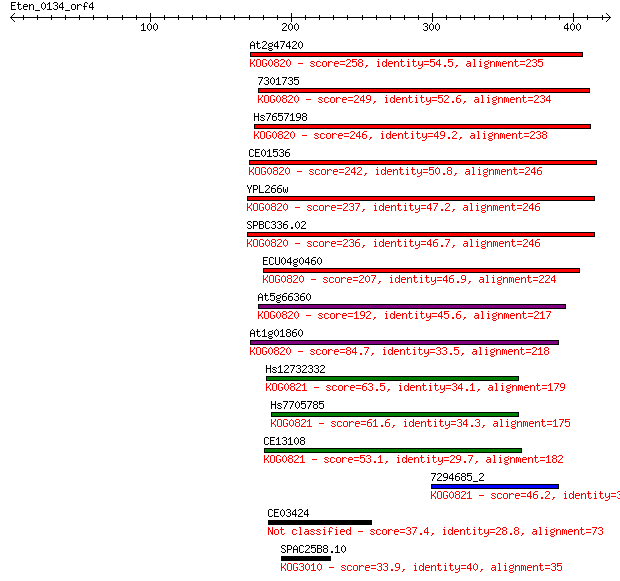
Query= Eten_0134_orf4
Length=425
Score E
Sequences producing significant alignments: (Bits) Value
At2g47420 258 2e-68
7301735 249 7e-66
Hs7657198 246 1e-64
CE01536 242 9e-64
YPL266w 237 4e-62
SPBC336.02 236 8e-62
ECU04g0460 207 2e-53
At5g66360 192 1e-48
At1g01860 84.7 3e-16
Hs12732332 63.5 8e-10
Hs7705785 61.6 3e-09
CE13108 53.1 1e-06
7294685_2 46.2 1e-04
CE03424 37.4 0.062
SPAC25B8.10 33.9 0.75
> At2g47420
Length=353
Score = 258 bits (658), Expect = 2e-68, Method: Compositional matrix adjust.
Identities = 128/236 (54%), Positives = 159/236 (67%), Gaps = 1/236 (0%)
Query 171 SSSSTAGFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAA 230
S+ G K GQH+LKNP ++D I+ A I S+DV+LEIGPGTGNLT +L +
Sbjct 19 SNHYQGGISFHKSKGQHILKNPLLVDSIVQKAGIKSTDVILEIGPGTGNLTKKLLEAGKE 78
Query 231 VRTLEIDARMAAEVYRRCKGLGFTN-LEVEMGDCLKKELGVFDVCAANLPYQISSPFLFK 289
V +E+D+RM E+ RR +G F+N L+V GD LK EL FD+C AN+PYQISSP FK
Sbjct 79 VIAVELDSRMVLELQRRFQGTPFSNRLKVIQGDVLKTELPRFDICVANIPYQISSPLTFK 138
Query 290 LAAHKHPFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPPP 349
L H FRCAV+M Q EF RL+A G YCRL+VN L+ VS + KV +N+FRPPP
Sbjct 139 LLFHPTSFRCAVIMYQREFAMRLVAQPGDNLYCRLSVNTQLYARVSHLLKVGKNNFRPPP 198
Query 350 KVDSVVVKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETNHK 405
KVDS VV+I P+R V+ EW+G LR+CF RK KT+ SIFKQ SVLS LE N K
Sbjct 199 KVDSSVVRIEPRRPGPQVNKKEWDGFLRVCFIRKNKTLGSIFKQKSVLSMLEKNFK 254
> 7301735
Length=306
Score = 249 bits (637), Expect = 7e-66, Method: Compositional matrix adjust.
Identities = 123/235 (52%), Positives = 159/235 (67%), Gaps = 1/235 (0%)
Query 177 GFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAAVRTLEI 236
G K +GQH+LKNP V+ ++ A + ++DVVLEIGPGTGN+TVR+ A V EI
Sbjct 20 GIVFNKDFGQHILKNPLVITTMLEKAALRATDVVLEIGPGTGNMTVRMLERAKKVIACEI 79
Query 237 DARMAAEVYRRCKGLGFT-NLEVEMGDCLKKELGVFDVCAANLPYQISSPFLFKLAAHKH 295
D R+AAE+ +R + L+V +GD LK EL FD+C AN+PYQISSP +FKL H+
Sbjct 80 DTRLAAELQKRVQATPLQPKLQVLIGDFLKAELPFFDLCIANVPYQISSPLIFKLLLHRP 139
Query 296 PFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPPPKVDSVV 355
FRCAVLM Q EF ERL+A G K YCRL++N L V + KV +N+FRPPPKV+S V
Sbjct 140 LFRCAVLMFQREFAERLVAKPGDKLYCRLSINTQLLARVDMLMKVGKNNFRPPPKVESSV 199
Query 356 VKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETNHKLFCSL 410
V++ PK PV+F EW+GL R+ F RK KT+ + FK SVL LE N+KL+ SL
Sbjct 200 VRLEPKNPPPPVNFTEWDGLTRIAFLRKNKTLAATFKVTSVLEMLEKNYKLYRSL 254
> Hs7657198
Length=313
Score = 246 bits (627), Expect = 1e-64, Method: Compositional matrix adjust.
Identities = 117/239 (48%), Positives = 160/239 (66%), Gaps = 1/239 (0%)
Query 174 STAGFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAAVRT 233
S G GQH+LKNP +++ II A + +DVVLE+GPGTGN+TV+L A V
Sbjct 24 SAGGLMFNTGIGQHILKNPLIINSIIDKAALRPTDVVLEVGPGTGNMTVKLLEKAKKVVA 83
Query 234 LEIDARMAAEVYRRCKGLGF-TNLEVEMGDCLKKELGVFDVCAANLPYQISSPFLFKLAA 292
E+D R+ AE+++R +G + L+V +GD LK +L FD C ANLPYQISSPF+FKL
Sbjct 84 CELDPRLVAELHKRVQGTPVASKLQVLVGDVLKTDLPFFDTCVANLPYQISSPFVFKLLL 143
Query 293 HKHPFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPPPKVD 352
H+ FRCA+LM Q EF RL+A G K YCRL++N L V + KV +N+FRPPPKV+
Sbjct 144 HRPFFRCAILMFQREFALRLVAKPGDKLYCRLSINTQLLARVDHLMKVGKNNFRPPPKVE 203
Query 353 SVVVKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETNHKLFCSLN 411
S VV+I PK P++F EW+GL+R+ F RK KT+ + FK +V LE N+++ CS++
Sbjct 204 SSVVRIEPKNPPPPINFQEWDGLVRITFVRKNKTLSAAFKSSAVQQLLEKNYRIHCSVH 262
> CE01536
Length=304
Score = 242 bits (618), Expect = 9e-64, Method: Compositional matrix adjust.
Identities = 125/248 (50%), Positives = 158/248 (63%), Gaps = 6/248 (2%)
Query 170 SSSSSTAGFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAA 229
SS+ + P GQH+LKNPGV++ I+ + + ++D VLE+GPGTGNLTV++ A
Sbjct 14 SSTGNVQSLPFNTDKGQHILKNPGVVNAIVEKSALKATDTVLEVGPGTGNLTVKMLEVAK 73
Query 230 AVRTLEIDARMAAEVYRRCKGLGFTN-LEVEMGDCLKKELGVFDVCAANLPYQISSPFLF 288
V EID RM AEV +R G N L+V GD +K E FDVC ANLPYQISSPF+
Sbjct 74 TVIACEIDPRMIAEVKKRVMGTPLQNKLQVNGGDVMKMEWPFFDVCVANLPYQISSPFVQ 133
Query 289 KLAAHKHPFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPP 348
KL H+ R AVLM Q+EF +RL+A G K+Y RL+VNV L V + KV R FRPP
Sbjct 134 KLLLHRPLPRYAVLMFQKEFADRLVARPGDKDYSRLSVNVQLLAKVEMLMKVKRTEFRPP 193
Query 349 PKVDSVVVKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETNHKLFC 408
PKVDS VV+I PK P EW GLLRLCF RK KT+ +IF+ +V+ +E N + C
Sbjct 194 PKVDSAVVRIAPKNPPPP----EWEGLLRLCFMRKNKTLMAIFRLSNVIEVIEDNFRKVC 249
Query 409 SL-NKKLP 415
S NK +P
Sbjct 250 SFKNKPIP 257
> YPL266w
Length=318
Score = 237 bits (604), Expect = 4e-62, Method: Compositional matrix adjust.
Identities = 116/247 (46%), Positives = 159/247 (64%), Gaps = 1/247 (0%)
Query 169 SSSSSSTAGFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASA 228
S+ ++ F GQH+LKNP V I+ A I SDVVLE+GPGTGNLTVR+ A
Sbjct 19 SAEKHLSSVFKFNTDLGQHILKNPLVAQGIVDKAQIRPSDVVLEVGPGTGNLTVRILEQA 78
Query 229 AAVRTLEIDARMAAEVYRRCKGLGFTN-LEVEMGDCLKKELGVFDVCAANLPYQISSPFL 287
V +E+D RMAAE+ +R +G LE+ +GD +K EL FD+C +N PYQISSP +
Sbjct 79 KNVVAVEMDPRMAAELTKRVRGTPVEKKLEIMLGDFMKTELPYFDICISNTPYQISSPLV 138
Query 288 FKLAAHKHPFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRP 347
FKL P R ++LM Q EF RLLA G YCRL+ NV ++ NV+ + KV +N+FRP
Sbjct 139 FKLINQPRPPRVSILMFQREFALRLLARPGDSLYCRLSANVQMWANVTHIMKVGKNNFRP 198
Query 348 PPKVDSVVVKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETNHKLF 407
PP+V+S VV++ K VD+ EW+GLLR+ F RK +TI + FK +V+ LE N+K F
Sbjct 199 PPQVESSVVRLEIKNPRPQVDYNEWDGLLRIVFVRKNRTISAGFKSTTVMDILEKNYKTF 258
Query 408 CSLNKKL 414
++N ++
Sbjct 259 LAMNNEM 265
> SPBC336.02
Length=307
Score = 236 bits (602), Expect = 8e-62, Method: Compositional matrix adjust.
Identities = 115/247 (46%), Positives = 159/247 (64%), Gaps = 1/247 (0%)
Query 169 SSSSSSTAGFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASA 228
S + F K +GQH+LKNP V I+ A++ SD VLE+GPGTGNLTVR+ A
Sbjct 13 SDAEVRNTVFKFNKDFGQHILKNPLVAQGIVDKADLKQSDTVLEVGPGTGNLTVRMLEKA 72
Query 229 AAVRTLEIDARMAAEVYRRCKGL-GFTNLEVEMGDCLKKELGVFDVCAANLPYQISSPFL 287
V +E+D RMAAE+ +R +G L+V +GD +K +L FDVC +N PYQISSP +
Sbjct 73 RKVIAVEMDPRMAAEITKRVQGTPKEKKLQVVLGDVIKTDLPYFDVCVSNTPYQISSPLV 132
Query 288 FKLAAHKHPFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRP 347
FKL + R A+LM Q EF RL+A G YCRL+ NV ++ +V + KV +N+FRP
Sbjct 133 FKLLQQRPAPRAAILMFQREFALRLVARPGDPLYCRLSANVQMWAHVKHIMKVGKNNFRP 192
Query 348 PPKVDSVVVKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETNHKLF 407
PP V+S VV+I PK P+ F EW+GLLR+ F RK KTI + FK S++ +E N++ +
Sbjct 193 PPLVESSVVRIEPKNPPPPLAFEEWDGLLRIVFLRKNKTIGACFKTSSIIEMVENNYRTW 252
Query 408 CSLNKKL 414
CS N+++
Sbjct 253 CSQNERM 259
> ECU04g0460
Length=277
Score = 207 bits (528), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 105/226 (46%), Positives = 148/226 (65%), Gaps = 4/226 (1%)
Query 180 LQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAAVRTLEIDAR 239
K++GQH+LKN GV+D II A I +D VLE+G GTGNLT++L A V EID R
Sbjct 6 FNKQHGQHILKNLGVIDTIIERARIKPTDTVLEVGGGTGNLTMKLLQKAKKVVCYEIDPR 65
Query 240 MAAEVYRRCKGL-GFTN-LEVEMGDCLKKELGVFDVCAANLPYQISSPFLFKLAAHKHPF 297
+AAE+ ++ G G ++ +GD LK + FD+C NLPYQISSPF+FKL K+ F
Sbjct 66 LAAELAKKVNGTPGMAQKFQLFVGDALKHDFPHFDLCITNLPYQISSPFVFKLL--KYDF 123
Query 298 RCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPPPKVDSVVVK 357
+CA +M Q+EF +RL+A GS YCRL+V + + V KV +NSF PPPKV+S V+
Sbjct 124 KCAFIMFQKEFSDRLVARPGSPEYCRLSVGAQILAQIDHVLKVSKNSFVPPPKVESSFVR 183
Query 358 ITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQPSVLSFLETN 403
I P+ P+D E++ L++CF RK KT+R+ FK S++S ++ +
Sbjct 184 IEPRIPRPPIDLGEFDRFLKICFLRKNKTLRANFKNSSLVSKIKND 229
> At5g66360
Length=380
Score = 192 bits (489), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 99/218 (45%), Positives = 133/218 (61%), Gaps = 1/218 (0%)
Query 177 GFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAAVRTLEI 236
G L K GQHLL N +LD I+ +++I +D VLEIGPGTGNLT++L +A V +E+
Sbjct 60 GLFLCKSKGQHLLTNTRILDSIVRSSDIRPTDTVLEIGPGTGNLTMKLLEAAQNVVAVEL 119
Query 237 DARMAAEVYRRCKGLGFTN-LEVEMGDCLKKELGVFDVCAANLPYQISSPFLFKLAAHKH 295
D RM + +R GF + L + D LK + FD+ AN+PY ISSP + KL +
Sbjct 120 DKRMVEILRKRVSDHGFADKLTIIQKDVLKTDFPHFDLVVANIPYNISSPLVAKLVYGSN 179
Query 296 PFRCAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPPPKVDSVV 355
FR A L+LQ+EF RLLA G ++ RL VNV L +V V V + F PPPKVDS V
Sbjct 180 TFRSATLLLQKEFSRRLLANPGDSDFNRLAVNVKLVADVKFVMDVSKREFVPPPKVDSSV 239
Query 356 VKITPKREVLPVDFYEWNGLLRLCFSRKRKTIRSIFKQ 393
++ITPK + V+ EW R CF +K KT+ S+F+Q
Sbjct 240 IRITPKEIIPDVNVQEWLAFTRTCFGKKNKTLGSMFRQ 277
> At1g01860
Length=343
Score = 84.7 bits (208), Expect = 3e-16, Method: Compositional matrix adjust.
Identities = 73/240 (30%), Positives = 110/240 (45%), Gaps = 26/240 (10%)
Query 171 SSSSTAGFPLQKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAA 230
S +S FP +K GQH + N + D++ +AA++ D VLEIGPGTG+LT L A
Sbjct 63 SLNSRGRFP-RKSLGQHYMLNSDINDQLASAADVKEGDFVLEIGPGTGSLTNVLINLGAT 121
Query 231 VRTLEIDARMAAEVYRRCKGLG-FTNLEVEMGDCLKKE--LGVFDV------------CA 275
V +E D M V R G F L+ + C + L + +
Sbjct 122 VLAIEKDPHMVDLVSERFAGSDKFKVLQEDFVKCHIRSHMLSILETRRLSHPDSALAKVV 181
Query 276 ANLPYQISSPFLFKLAAHKHPFRCAVLMLQEEFGERLLAAA-GSKNYCRLTVNVNLFLNV 334
+NLP+ IS+ + L F VL+LQ+E RL+ A + Y + + +N +
Sbjct 182 SNLPFNISTDVVKLLLPMGDIFSKVVLLLQDEAALRLVEPALRTSEYRPINILINFYSEP 241
Query 335 SRVCKVDRNSFRPPPKVDSVVVKITPKREVLPVDFYE------WNGLLRLCFSRKRKTIR 388
+V R +F P PKVD+ VV K P D+ + + L+ F+ KRK +R
Sbjct 242 EYNFRVPRENFFPQPKVDAAVVTFKLKH---PRDYPDVSSTKNFFSLVNSAFNGKRKMLR 298
> Hs12732332
Length=346
Score = 63.5 bits (153), Expect = 8e-10, Method: Compositional matrix adjust.
Identities = 61/202 (30%), Positives = 88/202 (43%), Gaps = 23/202 (11%)
Query 182 KKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVR-LCASAAAVRTLEIDARM 240
K+ Q+ L + + DKI+ A ++ V E+GPG G +T L A A + +E D R
Sbjct 31 KQLSQNFLLDLRLTDKIVRKAGNLTNAYVYEVGPGPGGITRSILNADVAELLVVEKDTRF 90
Query 241 ----------AAEVYRRCKGLGFT-NLEVEMGDCLKK--ELGVFDV-CAANLPYQISSPF 286
A R G T +E + LK+ E +V NLP+ +S+P
Sbjct 91 IPGLQMLSDAAPGKLRIVHGDVLTFKVEKAFSESLKRPWEDDPPNVHIIGNLPFSVSTPL 150
Query 287 LFK----LAAHKHPFRCA----VLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVC 338
+ K ++ PF L Q+E ERL A GSK RL+V NV +
Sbjct 151 IIKWLENISCRDGPFVYGRTQMTLTFQKEVAERLAANTGSKQRSRLSVMAQYLCNVRHIF 210
Query 339 KVDRNSFRPPPKVDSVVVKITP 360
+ +F P P+VD VV TP
Sbjct 211 TIPGQAFVPKPEVDVGVVHFTP 232
> Hs7705785
Length=346
Score = 61.6 bits (148), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 60/198 (30%), Positives = 86/198 (43%), Gaps = 23/198 (11%)
Query 186 QHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVR-LCASAAAVRTLEIDARM---- 240
Q+ L + + DKI+ A ++ V E+GPG G +T L A A + +E D R
Sbjct 35 QNFLLDLRLTDKIVRKAGNLTNAYVYEVGPGPGGITRSILNADVAELLVVEKDTRFIPGL 94
Query 241 ------AAEVYRRCKGLGFT-NLEVEMGDCLKK--ELGVFDV-CAANLPYQISSPFLFK- 289
A R G T +E + LK+ E +V NLP+ +S+P + K
Sbjct 95 QMLSDAAPGKLRIVHGDVLTFKVEKAFSESLKRPWEDDPPNVHIIGNLPFSVSTPLIIKW 154
Query 290 ---LAAHKHPFRCA----VLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDR 342
++ PF L Q+E ERL A GSK RL+V NV + +
Sbjct 155 LENISCRDGPFVYGRTQMTLTFQKEVAERLAANTGSKQRSRLSVMAQYLCNVRHIFTIPG 214
Query 343 NSFRPPPKVDSVVVKITP 360
+F P P+VD VV TP
Sbjct 215 QAFVPKPEVDVGVVHFTP 232
> CE13108
Length=313
Score = 53.1 bits (126), Expect = 1e-06, Method: Compositional matrix adjust.
Identities = 54/211 (25%), Positives = 85/211 (40%), Gaps = 30/211 (14%)
Query 181 QKKYGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLT-VRLCASAAAVRTLEIDAR 239
+K Q+ L + + KI A + D V+EIGPG G +T L A A+ + +EID R
Sbjct 25 KKILSQNYLMDMNITRKIAKHAKVIEKDWVIEIGPGPGGITRAILEAGASRLDVVEIDNR 84
Query 240 MAAEVYRRCKGLG---FTN----LEVEMGDCLKKE------LGVFDV------CAANLPY 280
+ + F + L E+GD K E + D NLP+
Sbjct 85 FIPPLQHLAEAADSRMFIHHQDALRTEIGDIWKNETARPESVDWHDSNLPAMHVIGNLPF 144
Query 281 QISSPFLFKLAAHKHPFRCAV---------LMLQEEFGERLLAAAGSKNYCRLTVNVNLF 331
I+SP + K +R V L Q E +RL + R+++
Sbjct 145 NIASPLIIKY-LRDMSYRRGVWQYGRVPLTLTFQLEVAKRLCSPIACDTRSRISIMSQYV 203
Query 332 LNVSRVCKVDRNSFRPPPKVDSVVVKITPKR 362
V ++ + F P P+VD VV+ P++
Sbjct 204 AEPKMVFQISGSCFVPRPQVDVGVVRFVPRK 234
> 7294685_2
Length=284
Score = 46.2 bits (108), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 29/91 (31%), Positives = 45/91 (49%), Gaps = 1/91 (1%)
Query 299 CAVLMLQEEFGERLLAAAGSKNYCRLTVNVNLFLNVSRVCKVDRNSFRPPPKVDSVVVKI 358
C L Q+E ER+ A G + CRL+V ++ + +F P P+VD VVK+
Sbjct 124 CMTLTFQQEVAERICAPVGGEQRCRLSVMSQVWTEPVMKFTIPGKAFVPKPQVDVGVVKL 183
Query 359 TP-KREVLPVDFYEWNGLLRLCFSRKRKTIR 388
P KR + F+ ++R FS ++K R
Sbjct 184 IPLKRPKTQLPFHLVERVVRHIFSMRQKYCR 214
> CE03424
Length=383
Score = 37.4 bits (85), Expect = 0.062, Method: Compositional matrix adjust.
Identities = 21/73 (28%), Positives = 36/73 (49%), Gaps = 3/73 (4%)
Query 184 YGQHLLKNPGVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCASAAAVRTLEIDARMAAE 243
YG+ +L+ G I+ +NSSDV +++G G G L + A + + + I+
Sbjct 157 YGETMLEQIG---SIVDELKLNSSDVFVDLGSGIGQLVCFVAAYSKVKKAVGIELSSVPA 213
Query 244 VYRRCKGLGFTNL 256
Y +G+ F NL
Sbjct 214 EYAVSQGMYFKNL 226
> SPAC25B8.10
Length=256
Score = 33.9 bits (76), Expect = 0.75, Method: Compositional matrix adjust.
Identities = 14/35 (40%), Positives = 21/35 (60%), Gaps = 0/35 (0%)
Query 193 GVLDKIIAAANINSSDVVLEIGPGTGNLTVRLCAS 227
G+ D I I+ + ++LE+G GTG T R+ AS
Sbjct 27 GITDWITDEFLIDETSIILELGAGTGKFTPRIIAS 61
Lambda K H
0.322 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 10979438924
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40