bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0124_orf1
Length=77
Score E
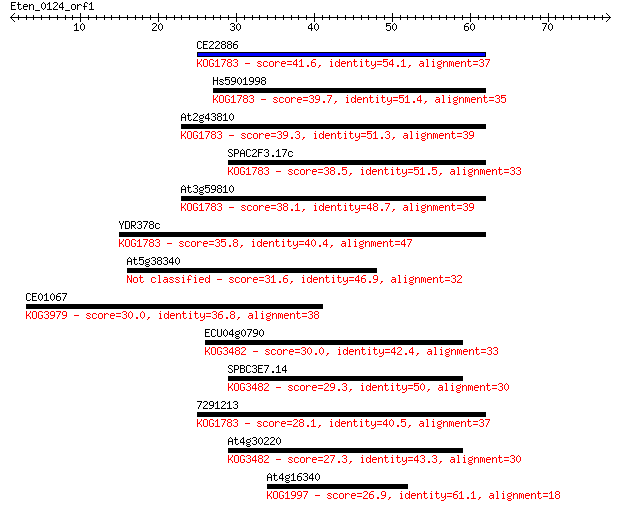
Sequences producing significant alignments: (Bits) Value
CE22886 41.6 4e-04
Hs5901998 39.7 0.001
At2g43810 39.3 0.002
SPAC2F3.17c 38.5 0.003
At3g59810 38.1 0.004
YDR378c 35.8 0.020
At5g38340 31.6 0.40
CE01067 30.0 0.97
ECU04g0790 30.0 1.1
SPBC3E7.14 29.3 2.0
7291213 28.1 4.0
At4g30220 27.3 7.8
At4g16340 26.9 8.4
> CE22886
Length=77
Score = 41.6 bits (96), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/37 (54%), Positives = 27/37 (72%), Gaps = 0/37 (0%)
Query 25 SGRKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
S R+ P+EFL++V+G V+VKLNSGV Y+G L G
Sbjct 2 SKRQNPAEFLKKVIGKPVVVKLNSGVDYRGILACLDG 38
> Hs5901998
Length=80
Score = 39.7 bits (91), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 27 RKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
++TPS+FL++++G V+VKLNSGV Y+G L G
Sbjct 5 KQTPSDFLKQIIGRPVVVKLNSGVDYRGVLACLDG 39
> At2g43810
Length=91
Score = 39.3 bits (90), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 20/39 (51%), Positives = 26/39 (66%), Gaps = 0/39 (0%)
Query 23 ASSGRKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
AS KTP++FL+ + G V+VKLNSGV Y+G L G
Sbjct 8 ASGTTKTPADFLKSIRGKPVVVKLNSGVDYRGILTCLDG 46
> SPAC2F3.17c
Length=75
Score = 38.5 bits (88), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 24/33 (72%), Gaps = 0/33 (0%)
Query 29 TPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
+P+EFL +V+G VL++L+SGV YKG L G
Sbjct 4 SPNEFLNKVIGKKVLIRLSSGVDYKGILSCLDG 36
> At3g59810
Length=91
Score = 38.1 bits (87), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 19/39 (48%), Positives = 25/39 (64%), Gaps = 0/39 (0%)
Query 23 ASSGRKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
S KTP++FL+ + G V+VKLNSGV Y+G L G
Sbjct 8 VSGTTKTPADFLKSIRGRPVVVKLNSGVDYRGTLTCLDG 46
> YDR378c
Length=123
Score = 35.8 bits (81), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 27/47 (57%), Gaps = 4/47 (8%)
Query 15 MTAKPPTSASSGRKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
M+ K T S +EFL ++G V VKL SG++Y GRL++ G
Sbjct 38 MSGKASTEGS----VTTEFLSDIIGKTVNVKLASGLLYSGRLESIDG 80
> At5g38340
Length=1059
Score = 31.6 bits (70), Expect = 0.40, Method: Composition-based stats.
Identities = 15/32 (46%), Positives = 18/32 (56%), Gaps = 0/32 (0%)
Query 16 TAKPPTSASSGRKTPSEFLQRVLGGNVLVKLN 47
T+ S GR+ PS F R GG+VLV LN
Sbjct 928 TSSSTCSILPGRRVPSNFTYRKTGGSVLVNLN 959
> CE01067
Length=278
Score = 30.0 bits (66), Expect = 0.97, Method: Composition-based stats.
Identities = 14/38 (36%), Positives = 19/38 (50%), Gaps = 0/38 (0%)
Query 3 HLLQDFSAHCEEMTAKPPTSASSGRKTPSEFLQRVLGG 40
H Q S HCE P SA+ TP +++ R+L G
Sbjct 54 HFDQATSTHCEVANWLPSISAAVSTYTPEKYIWRILIG 91
> ECU04g0790
Length=81
Score = 30.0 bits (66), Expect = 1.1, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 20/33 (60%), Gaps = 0/33 (0%)
Query 26 GRKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQA 58
R+ P +FLQR+ +V V L G +Y+G L A
Sbjct 10 ARENPKQFLQRMKNRSVRVALKWGQVYEGTLVA 42
> SPBC3E7.14
Length=78
Score = 29.3 bits (64), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 0/30 (0%)
Query 29 TPSEFLQRVLGGNVLVKLNSGVIYKGRLQA 58
P FLQ ++G VLV+L G YKG LQ+
Sbjct 7 NPKPFLQGLIGKPVLVRLKWGQEYKGTLQS 36
> 7291213
Length=79
Score = 28.1 bits (61), Expect = 4.0, Method: Compositional matrix adjust.
Identities = 15/37 (40%), Positives = 23/37 (62%), Gaps = 0/37 (0%)
Query 25 SGRKTPSEFLQRVLGGNVLVKLNSGVIYKGRLQAQHG 61
S ++ S+F+ ++ G V VKLN+GV Y+G L G
Sbjct 2 SRKEALSQFINQIHGRPVAVKLNNGVDYRGVLACLDG 38
> At4g30220
Length=88
Score = 27.3 bits (59), Expect = 7.8, Method: Compositional matrix adjust.
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 0/30 (0%)
Query 29 TPSEFLQRVLGGNVLVKLNSGVIYKGRLQA 58
P FL + G V+VKL G+ YKG L +
Sbjct 7 NPKPFLNNLTGKTVIVKLKWGMEYKGFLAS 36
> At4g16340
Length=1120
Score = 26.9 bits (58), Expect = 8.4, Method: Compositional matrix adjust.
Identities = 11/18 (61%), Positives = 16/18 (88%), Gaps = 0/18 (0%)
Query 34 LQRVLGGNVLVKLNSGVI 51
LQR+L G+V V++NSGV+
Sbjct 1025 LQRILQGSVAVQVNSGVL 1042
Lambda K H
0.316 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 1175087368
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40