bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
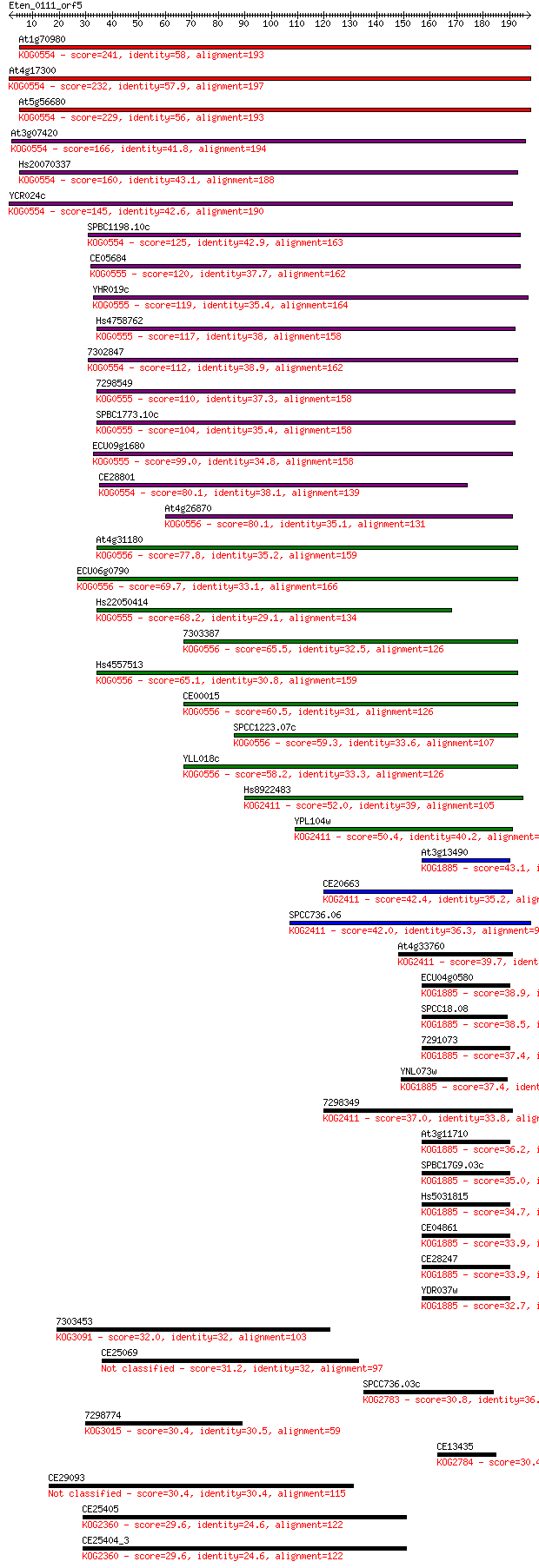
Query= Eten_0111_orf5
Length=197
Score E
Sequences producing significant alignments: (Bits) Value
At1g70980 241 9e-64
At4g17300 232 3e-61
At5g56680 229 3e-60
At3g07420 166 3e-41
Hs20070337 160 2e-39
YCR024c 145 6e-35
SPBC1198.10c 125 5e-29
CE05684 120 2e-27
YHR019c 119 5e-27
Hs4758762 117 2e-26
7302847 112 5e-25
7298549 110 1e-24
SPBC1773.10c 104 1e-22
ECU09g1680 99.0 6e-21
CE28801 80.1 2e-15
At4g26870 80.1 3e-15
At4g31180 77.8 1e-14
ECU06g0790 69.7 3e-12
Hs22050414 68.2 1e-11
7303387 65.5 7e-11
Hs4557513 65.1 8e-11
CE00015 60.5 2e-09
SPCC1223.07c 59.3 5e-09
YLL018c 58.2 1e-08
Hs8922483 52.0 8e-07
YPL104w 50.4 2e-06
At3g13490 43.1 3e-04
CE20663 42.4 6e-04
SPCC736.06 42.0 8e-04
At4g33760 39.7 0.004
ECU04g0580 38.9 0.006
SPCC18.08 38.5 0.008
7291073 37.4 0.020
YNL073w 37.4 0.020
7298349 37.0 0.026
At3g11710 36.2 0.044
SPBC17G9.03c 35.0 0.11
Hs5031815 34.7 0.11
CE04861 33.9 0.23
CE28247 33.9 0.24
YDR037w 32.7 0.48
7303453 32.0 0.74
CE25069 31.2 1.3
SPCC736.03c 30.8 2.0
7298774 30.4 2.4
CE13435 30.4 2.6
CE29093 30.4 2.7
CE25405 29.6 4.5
CE25404_3 29.6 4.5
> At1g70980
Length=571
Score = 241 bits (614), Expect = 9e-64, Method: Compositional matrix adjust.
Identities = 112/194 (57%), Positives = 147/194 (75%), Gaps = 1/194 (0%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
+D C +++ +KN +EG RL + A F+R+TYT+A+ RL++ A+G+V F KVEW
Sbjct 378 MDKCGDDMELMDKNVDEGCTKRLNMVAKASFKRVTYTEAIERLEKAVAQGKVVFDNKVEW 437
Query 65 GMDLGSEHERYLTE-QLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIG 123
G+DL SEHERYLTE + +KP+IV++YPK IKAFYM+ N+D KTV AMDVLVPK+GELIG
Sbjct 438 GIDLASEHERYLTEVEFDQKPIIVYNYPKGIKAFYMRLNDDEKTVAAMDVLVPKVGELIG 497
Query 124 GSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIR 183
GSQREE + + + I+E GL ++ Y WY +LR YGTV H GFGLGFER+I TGI+NIR
Sbjct 498 GSQREERYDVIKQRIEEMGLPMEPYEWYLDLRRYGTVKHCGFGLGFERMIQFATGIDNIR 557
Query 184 DTIPYPRYPGHAEF 197
D IP+PRYPG A+
Sbjct 558 DVIPFPRYPGKADL 571
> At4g17300
Length=567
Score = 232 bits (592), Expect = 3e-61, Method: Compositional matrix adjust.
Identities = 114/198 (57%), Positives = 146/198 (73%), Gaps = 5/198 (2%)
Query 1 VKSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQE 60
VK LDNC ++++F+ E+G+ RL ++ F +L YTDA+ L + KF
Sbjct 374 VKYVLDNCKEDMEFFDTWIEKGIIRRLSDVAEKEFLQLGYTDAIEILLKANK----KFDF 429
Query 61 KVEWGMDLGSEHERYLTEQ-LAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIG 119
V+WG+DL SEHERY+TE+ +PVI+ DYPK+IKAFYM++N+DGKTV AMD+LVP+IG
Sbjct 430 PVKWGLDLQSEHERYITEEAFGGRPVIIRDYPKEIKAFYMRENDDGKTVAAMDMLVPRIG 489
Query 120 ELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGI 179
ELIGGSQREE LE L+ + E LN +SYWWY +LR YG+VPHAGFGLGFERL+ VTGI
Sbjct 490 ELIGGSQREERLEVLEARLDELKLNKESYWWYLDLRRYGSVPHAGFGLGFERLVQFVTGI 549
Query 180 ENIRDTIPYPRYPGHAEF 197
+NIRD IP+PR P AEF
Sbjct 550 DNIRDVIPFPRTPASAEF 567
> At5g56680
Length=572
Score = 229 bits (583), Expect = 3e-60, Method: Compositional matrix adjust.
Identities = 108/193 (55%), Positives = 143/193 (74%), Gaps = 1/193 (0%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
L+ C +++ KN + G RL+ + + PF R+TYT A+ L+ A+G+ +F VEW
Sbjct 381 LEKCYADMELMAKNFDSGCIDRLKLVASTPFGRITYTKAIELLEEAVAKGK-EFDNNVEW 439
Query 65 GMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGG 124
G+DL SEHERYLTE L +KP+IV++YPK IKAFYM+ N+D KTV AMDVLVPK+GELIGG
Sbjct 440 GIDLASEHERYLTEVLFQKPLIVYNYPKGIKAFYMRLNDDEKTVAAMDVLVPKVGELIGG 499
Query 125 SQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRD 184
SQREE + + + I+E GL ++ Y WY +LR YGTV H GFGLGFER+I+ TG++NIRD
Sbjct 500 SQREERYDVIKKRIEEMGLPIEPYEWYLDLRRYGTVKHCGFGLGFERMILFATGLDNIRD 559
Query 185 TIPYPRYPGHAEF 197
IP+PRYPG A+
Sbjct 560 VIPFPRYPGKADL 572
> At3g07420
Length=638
Score = 166 bits (419), Expect = 3e-41, Method: Compositional matrix adjust.
Identities = 81/194 (41%), Positives = 120/194 (61%), Gaps = 3/194 (1%)
Query 2 KSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEK 61
K L+N ++ + K ++ + RLE ++ R +YT+ + LQ+ KF+ K
Sbjct 446 KYVLENRDEDMKFISKRVDKTITTRLEATASSSLLRFSYTEVISLLQKATT---TKFETK 502
Query 62 VEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGEL 121
EWG+ L +EH YLT+++ K PVIVH YPK IK FY++ N+D KTV A D++VPK+G +
Sbjct 503 PEWGVALTTEHLSYLTDEIYKGPVIVHTYPKAIKQFYVRLNDDKKTVAAFDLVVPKVGVV 562
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIEN 181
I GSQ EE E LD I E G + + WY +LR +GTV H+G L E++++ TG+ +
Sbjct 563 ITGSQNEERFEILDARIGESGFTREKFEWYLDLRRHGTVKHSGISLSMEQMLLYATGLPD 622
Query 182 IRDTIPYPRYPGHA 195
I+D IP+PR G A
Sbjct 623 IKDAIPFPRSWGKA 636
> Hs20070337
Length=250
Score = 160 bits (404), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 81/190 (42%), Positives = 115/190 (60%), Gaps = 7/190 (3%)
Query 5 LDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEW 64
L C +++ K G + RLE++L F ++YT+AV L++ F EW
Sbjct 61 LSKCPEDVELCHKFIAPGQKDRLEHMLKNNFLIISYTEAVEILKQASQ----NFTFTPEW 116
Query 65 GMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDG--KTVRAMDVLVPKIGELI 122
G DL +EHE+YL + PV V +YP +K FYM+ NEDG TV A+D+LVP +GEL
Sbjct 117 GADLRTEHEKYLVKHCGNIPVFVINYPLTLKPFYMRDNEDGPQHTVAAVDLLVPGVGELF 176
Query 123 GGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENI 182
GG REE L+ + GL + Y WY +LR +G+VPH GFG+GFER + + G++NI
Sbjct 177 GGGLREERYHFLEERLARSGLT-EVYQWYLDLRRFGSVPHGGFGMGFERYLQCILGVDNI 235
Query 183 RDTIPYPRYP 192
+D IP+PR+P
Sbjct 236 KDVIPFPRFP 245
> YCR024c
Length=492
Score = 145 bits (365), Expect = 6e-35, Method: Compositional matrix adjust.
Identities = 81/195 (41%), Positives = 118/195 (60%), Gaps = 11/195 (5%)
Query 1 VKSCLDNCSGEIDWFEKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQE 60
+ S +N S E+ ++ Q+ ++ R E+++ + +TYT+A+ L++ E F+
Sbjct 296 ISSQENNASSELSINQETQQ--IKTRWEDLINEKWHNITYTNAIEILKKRHNEVS-HFKY 352
Query 61 KVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKDIKAFYMKQNED-GKTVRAMDVLVPKIG 119
+ +WG L +EHE++L + K PV V DYP+ K FYMKQN TV D+LVP +G
Sbjct 353 EPKWGQPLQTEHEKFLAGEYFKSPVFVTDYPRLCKPFYMKQNSTPDDTVGCFDLLVPGMG 412
Query 120 ELIGGSQREEDLEKLDRMIKEKGLN----LDSYWWYRELRSYGTVPHAGFGLGFERLIML 175
E+IGGS RE+D +KL R +K +G+N LD WY LR G+ PH GFGLGFER I
Sbjct 413 EIIGGSLREDDYDKLCREMKARGMNRSGELD---WYVSLRKEGSAPHGGFGLGFERFISY 469
Query 176 VTGIENIRDTIPYPR 190
+ G NI+D IP+ R
Sbjct 470 LYGNHNIKDAIPFYR 484
> SPBC1198.10c
Length=441
Score = 125 bits (315), Expect = 5e-29, Method: Compositional matrix adjust.
Identities = 70/164 (42%), Positives = 98/164 (59%), Gaps = 13/164 (7%)
Query 31 LAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDY 90
L P++ +TY++A+ E + + ++ +WG DL SEHE+YL E L K PV V DY
Sbjct 286 LLKPWKCMTYSEAI----EELSAVKKTWKYPPKWGNDLSSEHEKYLCEILHKTPVFVTDY 341
Query 91 PKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
P+ IK FYMK + TV A+D+LVP++GEL GGS R++ L++ E W
Sbjct 342 PQKIKPFYMK-SSGPDTVAAVDLLVPQVGELAGGSLRKDHLDEYKTYPPE-------LQW 393
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTG-IENIRDTIPYPRYPG 193
Y +L Y PH GFGLG ERLI + G N+++TIP+PR G
Sbjct 394 YLDLMKYSNAPHGGFGLGIERLIAFLEGENTNVKETIPFPRSVG 437
> CE05684
Length=545
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 61/163 (37%), Positives = 101/163 (61%), Gaps = 4/163 (2%)
Query 32 AAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
A PF+R+ Y +A+ LQ+ + R + EK +G D+ ER +T+ + P++++ +P
Sbjct 382 ARPFKRMPYKEAIEWLQKNDV--RNEMGEKFVYGEDIAEAAERRMTDTIGV-PILLNRFP 438
Query 92 KDIKAFYM-KQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
IKAFYM + +D + ++D+L+P +GE++GGS R ++L ++ GL+ +Y+W
Sbjct 439 HGIKAFYMPRCADDNELTESVDLLMPGVGEIVGGSMRIWKEDQLLAAFEKGGLDSKNYYW 498
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPG 193
Y + R YG+VPH G+GLG ER I +T +IRD YPR+ G
Sbjct 499 YMDQRKYGSVPHGGYGLGLERFICWLTDTNHIRDVCLYPRFVG 541
> YHR019c
Length=554
Score = 119 bits (297), Expect = 5e-27, Method: Compositional matrix adjust.
Identities = 58/165 (35%), Positives = 100/165 (60%), Gaps = 4/165 (2%)
Query 33 APFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPK 92
APF RL Y DA+ L + + E ++G D+ ER +T+ + P+ + +P
Sbjct 392 APFMRLQYKDAITWLNEHDIKNEEG--EDFKFGDDIAEAAERKMTDTIGV-PIFLTRFPV 448
Query 93 DIKAFYMKQ-NEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWY 151
+IK+FYMK+ ++D + ++DVL+P +GE+ GGS R +D+++L K +G++ D+Y+W+
Sbjct 449 EIKSFYMKRCSDDPRVTESVDVLMPNVGEITGGSMRIDDMDELMAGFKREGIDTDAYYWF 508
Query 152 RELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPGHAE 196
+ R YGT PH G+G+G ER++ + +RD YPR+ G +
Sbjct 509 IDQRKYGTCPHGGYGIGTERILAWLCDRFTVRDCSLYPRFSGRCK 553
> Hs4758762
Length=548
Score = 117 bits (292), Expect = 2e-26, Method: Compositional matrix adjust.
Identities = 60/161 (37%), Positives = 101/161 (62%), Gaps = 8/161 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKV--EWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
PF+R+ Y+DA++ L+ E VK ++ E+G D+ ER +T+ + +P+++ +P
Sbjct 387 PFKRMNYSDAIVWLK----EHDVKKEDGTFYEFGEDIPEAPERLMTDTI-NEPILLCRFP 441
Query 92 KDIKAFYMKQN-EDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
+IK+FYM++ ED + ++DVL+P +GE++GGS R D E++ K +G++ Y+W
Sbjct 442 VEIKSFYMQRCPEDSRLTESVDVLMPNVGEIVGGSMRIFDSEEILAGYKREGIDPTPYYW 501
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
Y + R YGT PH G+GLG ER + + +IRD YPR+
Sbjct 502 YTDQRKYGTCPHGGYGLGLERFLTWILNRYHIRDVCLYPRF 542
> 7302847
Length=460
Score = 112 bits (280), Expect = 5e-25, Method: Compositional matrix adjust.
Identities = 63/163 (38%), Positives = 88/163 (53%), Gaps = 10/163 (6%)
Query 31 LAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDY 90
L P++ ++Y +A+ LQ E + + + + E +L PV V D+
Sbjct 302 LQVPWKIMSYDEALQVLQ----ENKDSLKTPISLNEGFSKDQELFLVAHCGA-PVFVVDW 356
Query 91 PKDIKAFYMKQNEDGKT-VRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYW 149
P K FYMK + V A+D+L+P +GEL GGS RE D ++K
Sbjct 357 PAQQKPFYMKVSRSNPNRVHALDLLMPAVGELCGGSLRESD----PAILKSHPHLPKDLE 412
Query 150 WYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
WY +LR YG VP GFG+GFER + LVTG++NIRD IP+PRYP
Sbjct 413 WYVDLRKYGGVPTGGFGMGFERYLQLVTGVKNIRDVIPFPRYP 455
> 7298549
Length=558
Score = 110 bits (276), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 59/161 (36%), Positives = 98/161 (60%), Gaps = 8/161 (4%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKV--EWGMDLGSEHERYLTEQLAKKPVIVHDYP 91
PF R+ Y DA+ L+ E V + E+G D+ ER +T+ + +P+++ +P
Sbjct 397 PFRRMNYEDAIKWLK----ENNVTKDDGTFYEFGEDIPEAPERKMTDTI-NEPIMLCRFP 451
Query 92 KDIKAFYMKQ-NEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWW 150
+IK+FYM++ ED + ++DVL+P +GE++GGS R D E+L + K +G++ Y+W
Sbjct 452 AEIKSFYMQRCKEDQRLTESVDVLLPNVGEIVGGSMRIYDSEELLKGYKREGIDPQPYYW 511
Query 151 YRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
Y + R YGT+PH G+GLG ER + + +IR+ YPR+
Sbjct 512 YTDQRVYGTMPHGGYGLGLERFLCWLLNRYHIREVCLYPRF 552
> SPBC1773.10c
Length=568
Score = 104 bits (259), Expect = 1e-22, Method: Compositional matrix adjust.
Identities = 56/162 (34%), Positives = 93/162 (57%), Gaps = 10/162 (6%)
Query 34 PFERLTYTDAVIRLQREE---AEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDY 90
PF RL+Y DA+ L EG E+ ++G D+ ER +T+Q+ +P+ + +
Sbjct 407 PFMRLSYEDAIKYLNEHNILTPEG-----EQHKFGDDIAEAAERKMTDQI-NRPIFLTYF 460
Query 91 PKDIKAFYMKQNED-GKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYW 149
P +IK+FYMK+ D + ++D L+P +GE++GGS R D+++L K +G++ Y+
Sbjct 461 PLEIKSFYMKRVVDRPELTESVDCLMPNVGEIVGGSMRISDIQELLAAYKREGIDPAPYY 520
Query 150 WYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRY 191
W+ E R YGT H G GLG ER + + +R+ +PR+
Sbjct 521 WFTEQRKYGTTEHGGCGLGLERFLAWLCDRYTVRECCLFPRF 562
> ECU09g1680
Length=502
Score = 99.0 bits (245), Expect = 6e-21, Method: Compositional matrix adjust.
Identities = 55/162 (33%), Positives = 92/162 (56%), Gaps = 8/162 (4%)
Query 33 APFERLTYTDAVIRLQRE--EAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKK-PVIVHD 89
PF+R+ Y++A+ L+ + + E F E G D+ ERYL E + PV +
Sbjct 338 TPFKRIKYSEAIELLKSKGYKKEDNTDF----ELGDDIPDAAERYLVEVVGDGCPVFLTH 393
Query 90 YPKDIKAFYMKQNEDGKTV-RAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSY 148
+ K FYM+++E+ K + + D+L P IGE++GGS R++ E L + + +N+D Y
Sbjct 394 FLVGHKPFYMRKDENDKGLTESTDLLFPGIGEILGGSMRQDTYEDLIEGFRRENINIDPY 453
Query 149 WWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
+WY ++ +G H G+GLGFER +M + E++ + YPR
Sbjct 454 YWYLDMARFGPCKHGGYGLGFERFLMGLMRYESVDEATLYPR 495
> CE28801
Length=448
Score = 80.1 bits (196), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 53/141 (37%), Positives = 76/141 (53%), Gaps = 21/141 (14%)
Query 35 FERLTYTDAV--IRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPK 92
F R+ Y +AV + L+ + + F +K E +DL H+ + P+ V YP
Sbjct 273 FPRIRYDEAVHLLALKGSKVTAKSGFSKKNE--LDLVKLHDDH--------PIFVTHYPV 322
Query 93 DIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYR 152
K FYM + DG+ + D+LVP IGEL GGS RE+ E+L R +G +D WY
Sbjct 323 GQKPFYMAR--DGQLTLSFDLLVPGIGELAGGSLREQSAEELQR----RGCGID---WYL 373
Query 153 ELRSYGTVPHAGFGLGFERLI 173
E+R G P GFG+GFER++
Sbjct 374 EMRRRGQPPTGGFGIGFERML 394
> At4g26870
Length=504
Score = 80.1 bits (196), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 46/136 (33%), Positives = 76/136 (55%), Gaps = 6/136 (4%)
Query 60 EKVEWGMDLGSEHERYLTEQLAKKP----VIVHDYPKDIKAFY-MKQNEDGKTVRAMDVL 114
E+V+ DL +E ER L + + +K ++H YP ++ FY M D + DV
Sbjct 363 EEVDPLGDLNTESERKLGQLVLEKYKTEFYMLHRYPSAVRPFYTMPYENDSNYSNSFDVF 422
Query 115 VPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIM 174
+ + E++ G+QR D E L++ +E G+++ + Y + YG PH GFG+G ER++M
Sbjct 423 I-RGEEIMSGAQRIHDPELLEKRARECGIDVKTISTYIDAFRYGAPPHGGFGVGLERVVM 481
Query 175 LVTGIENIRDTIPYPR 190
L+ + NIR T +PR
Sbjct 482 LLCALNNIRKTSLFPR 497
> At4g31180
Length=558
Score = 77.8 bits (190), Expect = 1e-14, Method: Compositional matrix adjust.
Identities = 56/170 (32%), Positives = 84/170 (49%), Gaps = 17/170 (10%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGM------DLGSEHERYLTEQLAKKP--- 84
PFE L + +RL EE +K E G+ DL +E ER L + + +K
Sbjct 390 PFEPLKFLPKTLRLTFEEGVQMLK-----EAGVEVDPLGDLNTESERKLGQLVLEKYNTE 444
Query 85 -VIVHDYPKDIKAFYMKQNEDGKT-VRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKG 142
I+H YPK ++ FY D + DV + + E+I G+QR E L++ E G
Sbjct 445 FYILHRYPKAVRPFYTMTCADNPLYSNSFDVFI-RGEEIISGAQRVHIPEVLEQRAGECG 503
Query 143 LNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
+++ + Y + YG H GFG+G ER++ML + NIR T +PR P
Sbjct 504 IDVKTISTYIDSFRYGAPLHGGFGVGLERVVMLFCALNNIRKTSLFPRDP 553
> ECU06g0790
Length=473
Score = 69.7 bits (169), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 55/174 (31%), Positives = 85/174 (48%), Gaps = 11/174 (6%)
Query 27 LENILA-APFERLTYT-DAVIRLQREEAEGRVKFQEKVEWGM--DLGSEHERYLTEQLAK 82
LE I A FE L Y D V+ RE + + E VE G D SE E+ L + +
Sbjct 298 LETIRAFHAFEDLKYRRDPVVLTHRECVD--LLLNEGVEMGYEDDFNSESEKKLGSVVRR 355
Query 83 KP----VIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMI 138
++ DYP + FY ++E+ R+ D ++ + E++ G+QR + L + +
Sbjct 356 MHGVDIFVIKDYPISTRPFYTYRDEEKGITRSYDFIL-RGQEILSGAQRVSIYKDLVKYV 414
Query 139 KEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
+E G++ S Y E YG PH G G+G ERL+ G+ +IR +PR P
Sbjct 415 EEHGISPSSLGGYLESFKYGAPPHGGCGIGLERLMKAYFGMGDIRCFSLFPRDP 468
> Hs22050414
Length=153
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 39/135 (28%), Positives = 75/135 (55%), Gaps = 4/135 (2%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVHDYPKD 93
PF+R+ Y+D ++ L+ E + + E+G D+ ER T+ + + + + +P +
Sbjct 19 PFKRINYSDTIVWLK--EHNIKKEHGTFYEFGEDIPEAPERVKTDTINEL-IFLCQFPLE 75
Query 94 IKAFYMKQN-EDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYR 152
IK+FY++Q ED ++D+L+ +GE +GGS D E++ + K ++ ++WY
Sbjct 76 IKSFYIQQCPEDSHLTESVDLLMSNVGENVGGSIHTCDGEEMLEGYEMKEIDPTPHYWYV 135
Query 153 ELRSYGTVPHAGFGL 167
+ R Y + PH +GL
Sbjct 136 DQRKYSSCPHGRYGL 150
> 7303387
Length=531
Score = 65.5 bits (158), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 41/130 (31%), Positives = 63/130 (48%), Gaps = 4/130 (3%)
Query 67 DLGSEHERYLTEQLAKKP----VIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELI 122
DL + +E+ L + K I+ +P I+ FY + + + + E++
Sbjct 397 DLSTPNEKLLGRLVKAKYDTDFYILDKFPLAIRPFYTMPDPNNPVYSNSYDMFMRGEEIL 456
Query 123 GGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENI 182
G+QR D E L K G++ Y E YG PHAG G+G ER++ML G++NI
Sbjct 457 SGAQRIHDPEYLIERAKHHGIDTSKIAAYIESFRYGCPPHAGGGIGMERVVMLYLGLDNI 516
Query 183 RDTIPYPRYP 192
R T +PR P
Sbjct 517 RKTSMFPRDP 526
> Hs4557513
Length=500
Score = 65.1 bits (157), Expect = 8e-11, Method: Compositional matrix adjust.
Identities = 49/166 (29%), Positives = 83/166 (50%), Gaps = 9/166 (5%)
Query 34 PFERLTYTDAVIRLQREEAEGRVKFQEKVEWGM--DLGSEHERYLTEQLAKKP----VIV 87
P E + + +RL+ EA ++ + VE G DL + +E+ L + +K I+
Sbjct 332 PCEPFKFLEPTLRLEYCEALAMLR-EAGVEMGDEDDLSTPNEKLLGHLVKEKYDTDFYIL 390
Query 88 HDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLD 146
YP ++ FY M + K ++ D+ + + E++ G+QR D + L G +L+
Sbjct 391 DKYPLAVRPFYTMPDPRNPKQSKSYDMFM-RGEEILSGAQRIHDPQLLTERALHHGNDLE 449
Query 147 SYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
Y + +G PHAG G+G ER+ ML G+ N+R T +PR P
Sbjct 450 KIKAYIDSFRFGAPPHAGGGIGLERVTMLFLGLHNVRQTSMFPRDP 495
> CE00015
Length=531
Score = 60.5 bits (145), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 39/131 (29%), Positives = 65/131 (49%), Gaps = 6/131 (4%)
Query 67 DLGSEHERYL----TEQLAKKPVIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGEL 121
DL + E++L E+ + ++ +P ++ FY M D + + D+ + + E+
Sbjct 397 DLSTPVEKFLGKLVKEKYSTDFYVLDKFPLSVRPFYTMPDAHDERYSNSYDMFM-RGEEI 455
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIEN 181
+ G+QR D + L K ++L Y + YG PHAG G+G ER+ ML G+ N
Sbjct 456 LSGAQRIHDADMLVERAKHHQVDLAKIQSYIDSFKYGCPPHAGGGIGLERVTMLFLGLHN 515
Query 182 IRDTIPYPRYP 192
IR +PR P
Sbjct 516 IRLASLFPRDP 526
> SPCC1223.07c
Length=580
Score = 59.3 bits (142), Expect = 5e-09, Method: Compositional matrix adjust.
Identities = 36/109 (33%), Positives = 56/109 (51%), Gaps = 3/109 (2%)
Query 86 IVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLN 144
++ YP ++ FY M ED + + D + K E++ G+QR D E L +K G++
Sbjct 468 VIDKYPSSVRPFYTMPDPEDPRYSNSYDFFM-KGQEIMSGAQRIHDPELLVERMKALGVS 526
Query 145 LD-SYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPRYP 192
D Y + + G PHAG G+G ER++M + NIR +PR P
Sbjct 527 PDVGLQQYIDAFAIGCPPHAGGGIGLERVVMFYLNLPNIRLASSFPRDP 575
> YLL018c
Length=557
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 42/133 (31%), Positives = 69/133 (51%), Gaps = 8/133 (6%)
Query 67 DLGSEHERYLTEQLAKKP----VIVHDYPKDIKAFY-MKQNEDGKTVRAMDVLVPKIGEL 121
DL +E+E++L + + K I+ +P +I+ FY M + K + D + + E+
Sbjct 421 DLSTENEKFLGKLVRDKYDTDFYILDKFPLEIRPFYTMPDPANPKYSNSYDFFM-RGEEI 479
Query 122 IGGSQREEDLEKLDRMIKEKGLNLDSYWW--YRELRSYGTVPHAGFGLGFERLIMLVTGI 179
+ G+QR D L +K GL+ + Y + SYG PHAG G+G ER++M +
Sbjct 480 LSGAQRIHDHALLQERMKAHGLSPEDPGLKDYCDGFSYGCPPHAGGGIGLERVVMFYLDL 539
Query 180 ENIRDTIPYPRYP 192
+NIR +PR P
Sbjct 540 KNIRRASLFPRDP 552
> Hs8922483
Length=463
Score = 52.0 bits (123), Expect = 8e-07, Method: Compositional matrix adjust.
Identities = 41/108 (37%), Positives = 51/108 (47%), Gaps = 7/108 (6%)
Query 90 YPKDIKAFYMKQNEDGKTVRAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLDSYW 149
+P DI Y E K LV E+ GGS R + E L R I L D
Sbjct 326 HPSDIHLLY---TEPKKARSQHYDLVLNGNEIGGGSIRIHNAE-LQRYILATLLKEDVKM 381
Query 150 WYRELRS--YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR-YPGH 194
L++ YG PH G LG +RLI LVTG +IRD I +P+ + GH
Sbjct 382 LSHLLQALDYGAPPHGGIALGLDRLICLVTGSPSIRDVIAFPKSFRGH 429
> YPL104w
Length=658
Score = 50.4 bits (119), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 33/86 (38%), Positives = 49/86 (56%), Gaps = 6/86 (6%)
Query 109 RAMDVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNLD-SYWWYRELRS---YGTVPHAG 164
R D++V + EL GGS R D +L I E L +D +Y + L + GT PHAG
Sbjct 543 RHYDLVVNGV-ELGGGSTRIHD-PRLQDYIFEDILKIDNAYELFGHLLNAFDMGTPPHAG 600
Query 165 FGLGFERLIMLVTGIENIRDTIPYPR 190
F +GF+R+ ++ E+IRD I +P+
Sbjct 601 FAIGFDRMCAMICETESIRDVIAFPK 626
> At3g13490
Length=602
Score = 43.1 bits (100), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 17/33 (51%), Positives = 23/33 (69%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P +G GLG +RL+ML+T +IRD I +P
Sbjct 564 YGMPPASGMGLGIDRLVMLLTNSASIRDVIAFP 596
> CE20663
Length=569
Score = 42.4 bits (98), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 25/75 (33%), Positives = 37/75 (49%), Gaps = 4/75 (5%)
Query 120 ELIGGSQREEDLEKLDRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGI 179
E+ GGS R E+ E ++K G D S+G PH GF LG +R + ++T
Sbjct 462 EMGGGSIRIENSEIQRHVLKVLGEPTDEMEHLLNALSHGAPPHGGFALGLDRFVAMLTSD 521
Query 180 EN----IRDTIPYPR 190
N +RD I +P+
Sbjct 522 GNPLTPVRDVIAFPK 536
> SPCC736.06
Length=611
Score = 42.0 bits (97), Expect = 8e-04, Method: Compositional matrix adjust.
Identities = 33/98 (33%), Positives = 50/98 (51%), Gaps = 9/98 (9%)
Query 107 TVRAM--DVLVPKIGELIGGSQREEDLEKLDRMIKEKGLNL--DSYWWYREL---RSYGT 159
+VR + D++V I EL GGS R + + + R + + L L + Y + L S G
Sbjct 488 SVRGLHYDIVVNGI-ELGGGSIRIHNPD-IQRFVLKDVLKLPENRYATFEHLIRVLSSGC 545
Query 160 VPHAGFGLGFERLIMLVTGIENIRDTIPYPRYPGHAEF 197
PH G LGF+RL L+T IR+ I +P+ A+
Sbjct 546 PPHGGIALGFDRLAALLTNAPGIREVIAFPKTSSGADL 583
> At4g33760
Length=636
Score = 39.7 bits (91), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 0/43 (0%)
Query 148 YWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPYPR 190
+ + E G PH G G +R++M++ G +IRD I +P+
Sbjct 566 FGYLLEALDMGAPPHGGIAYGLDRMVMMLGGASSIRDVIAFPK 608
> ECU04g0580
Length=440
Score = 38.9 bits (89), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P G+G+G +RL+M +T NIRD I +P
Sbjct 403 YGLPPTGGWGIGIDRLVMYLTDAANIRDVIFFP 435
> SPCC18.08
Length=531
Score = 38.5 bits (88), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 15/32 (46%), Positives = 23/32 (71%), Gaps = 0/32 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPY 188
YG P AG+G+G +RL+ML+TG I + +P+
Sbjct 494 YGLPPTAGWGMGVDRLVMLMTGQSKISEILPF 525
> 7291073
Length=572
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 14/33 (42%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P GFG+G +RL M +T NI++ + +P
Sbjct 520 YGLPPTGGFGMGIDRLAMFLTDSNNIKEVLLFP 552
> YNL073w
Length=576
Score = 37.4 bits (85), Expect = 0.020, Method: Compositional matrix adjust.
Identities = 16/40 (40%), Positives = 22/40 (55%), Gaps = 0/40 (0%)
Query 149 WWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIRDTIPY 188
+ Y E YG P GFGLG +RL ML + I + +P+
Sbjct 528 YQYVETMKYGMPPVGGFGLGIDRLCMLFCDKKRIEEVLPF 567
> 7298349
Length=1052
Score = 37.0 bits (84), Expect = 0.026, Method: Compositional matrix adjust.
Identities = 24/73 (32%), Positives = 35/73 (47%), Gaps = 3/73 (4%)
Query 120 ELIGGSQREEDLEKLDRMIKEKGLNL--DSYWWYRELRSYGTVPHAGFGLGFERLIMLVT 177
E+ GGS R D + + I E+ L + D G PH G LG +RLI ++
Sbjct 856 EVGGGSIRIHDRD-MQHFILEQILKIPHDHLSHLLSALESGCPPHGGIALGLDRLIAILC 914
Query 178 GIENIRDTIPYPR 190
+IRD I +P+
Sbjct 915 RARSIRDVIAFPK 927
> At3g11710
Length=626
Score = 36.2 bits (82), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P G+GLG +RL ML+T NI++ + +P
Sbjct 573 YGLAPTGGWGLGIDRLSMLLTDSLNIKEVLFFP 605
> SPBC17G9.03c
Length=591
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 21/33 (63%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P G+G+G +RL+M +T IR+ + +P
Sbjct 543 YGLPPTGGWGMGVDRLVMFLTDSNTIREVLLFP 575
> Hs5031815
Length=597
Score = 34.7 bits (78), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P AG+G+G +R+ M +T NI++ + +P
Sbjct 539 YGLPPTAGWGMGIDRVAMFLTDSNNIKEVLLFP 571
> CE04861
Length=572
Score = 33.9 bits (76), Expect = 0.23, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P G+G+G +RL M++T NI++ + +P
Sbjct 519 YGLPPTGGWGMGIDRLSMILTDNNNIKEVLLFP 551
> CE28247
Length=596
Score = 33.9 bits (76), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 22/33 (66%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P G+G+G +RL M++T NI++ + +P
Sbjct 543 YGLPPTGGWGMGIDRLSMILTDNNNIKEVLLFP 575
> YDR037w
Length=591
Score = 32.7 bits (73), Expect = 0.48, Method: Compositional matrix adjust.
Identities = 13/33 (39%), Positives = 19/33 (57%), Gaps = 0/33 (0%)
Query 157 YGTVPHAGFGLGFERLIMLVTGIENIRDTIPYP 189
YG P G+G G +RL M +T IR+ + +P
Sbjct 542 YGLPPTGGWGCGIDRLAMFLTDSNTIREVLLFP 574
> 7303453
Length=610
Score = 32.0 bits (71), Expect = 0.74, Method: Composition-based stats.
Identities = 33/113 (29%), Positives = 53/113 (46%), Gaps = 30/113 (26%)
Query 19 QEEGLRARLENILA---APFE---RLTYTDAVIRLQREE--AEGRVKFQEKVEWGMDLGS 70
+EE LR +L+N+LA AP + RL+ + +R+QR + A G E+ +D +
Sbjct 515 EEEALRTKLQNMLAVVSAPTQFKGRLSELLSQMRMQRNQFAANG------GAEYALDKEA 568
Query 71 EHE--RYLTEQLAKKPVIVHDYPKDIKAFYMKQNEDGKTVRAMDVLVPKIGEL 121
E E +LT Q V+ KD+ RA+DV++ + EL
Sbjct 569 EDEMKTFLTMQQRAMEVLSDTVNKDL--------------RALDVIIKGLPEL 607
> CE25069
Length=1327
Score = 31.2 bits (69), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 31/111 (27%), Positives = 53/111 (47%), Gaps = 14/111 (12%)
Query 36 ERLTYTDAVIRLQRE-EAEGRVKFQEKVEWGMDLGSE--------HERYLTEQLAKKPVI 86
E L D++++ E +A R+K E + W L E + YLTE ++ PV+
Sbjct 129 EDLIIEDSIVKTDEEKQAVRRLKINEFLSWFTRLLPEQFKNFEFTNPNYLTESISDSPVV 188
Query 87 VHDYPKDI-KAFYMKQNEDGKTVR----AMDVLVPKIGELIGGSQREEDLE 132
D K+I K+F ++ +G + + DVLV I + + EED++
Sbjct 189 NVDKCKEIVKSFKESESLEGLSQKYELIDEDVLVAAICIGVLDTNNEEDVD 239
> SPCC736.03c
Length=429
Score = 30.8 bits (68), Expect = 2.0, Method: Compositional matrix adjust.
Identities = 18/49 (36%), Positives = 24/49 (48%), Gaps = 14/49 (28%)
Query 135 DRMIKEKGLNLDSYWWYRELRSYGTVPHAGFGLGFERLIMLVTGIENIR 183
DR++K GLN W FG+G ERL M++ GI +IR
Sbjct 280 DRLLKGAGLNNYIGW--------------AFGIGLERLAMILYGIPDIR 314
> 7298774
Length=279
Score = 30.4 bits (67), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 18/59 (30%), Positives = 29/59 (49%), Gaps = 3/59 (5%)
Query 30 ILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQLAKKPVIVH 88
+ A E++ + + L R E E ++KF +VEW DL + L +LA + VH
Sbjct 210 LACAVIEQMLTVEKAVALARLEEEYQLKFWGRVEWAHDLSQQE---LQARLAAAVLFVH 265
> CE13435
Length=552
Score = 30.4 bits (67), Expect = 2.6, Method: Compositional matrix adjust.
Identities = 13/22 (59%), Positives = 15/22 (68%), Gaps = 0/22 (0%)
Query 163 AGFGLGFERLIMLVTGIENIRD 184
AG+GL ER M+ GI NIRD
Sbjct 510 AGYGLSLERPTMIKYGINNIRD 531
> CE29093
Length=772
Score = 30.4 bits (67), Expect = 2.7, Method: Composition-based stats.
Identities = 35/127 (27%), Positives = 61/127 (48%), Gaps = 22/127 (17%)
Query 16 EKNQEEGLRARLENILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERY 75
+K QEE +RA+LE A E+ R +REE + R++ K+E+ + E +R
Sbjct 482 QKKQEEEIRAKLEAKKKAEQEKE-------RAKREE-QRRIEEAAKLEYRRRI-EESQRL 532
Query 76 LTEQLAKKPVIVHDYPKDIKAFY------MKQNEDGKTV------RAMDVLVPKIGELIG 123
E+LA + +V D +D+ F +K + D K + R ++ + K E +
Sbjct 533 EAERLALEATMV-DEDEDVAKFIRNLEERIKDSRDSKNLNGALVGRQINQSLAKNAENLT 591
Query 124 GSQREED 130
+Q +ED
Sbjct 592 SAQTDED 598
> CE25405
Length=469
Score = 29.6 bits (65), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 30/130 (23%), Positives = 54/130 (41%), Gaps = 20/130 (15%)
Query 29 NILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQL-AKKPVIV 87
+ AAP + ++ A++ E +G+V W MD ++ + + L A K I
Sbjct 240 DTCAAPGMKTSHAAAIM-----ENQGKV-------WAMDRAADRVATMKQLLDASKVAIA 287
Query 88 HDYPKDIKAFYMKQNEDGKTVRAM-------DVLVPKIGELIGGSQREEDLEKLDRMIKE 140
+ D + + K A+ +V ++ E+ GG+ +E LEKL +
Sbjct 288 SSFCGDFLKTDVTDKKFSKVKFAIVDPPCSGSGIVKRMDEITGGNAEKERLEKLKNLQVH 347
Query 141 KGLNLDSYWW 150
K L+L W
Sbjct 348 KNLDLLKMCW 357
> CE25404_3
Length=436
Score = 29.6 bits (65), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 30/130 (23%), Positives = 54/130 (41%), Gaps = 20/130 (15%)
Query 29 NILAAPFERLTYTDAVIRLQREEAEGRVKFQEKVEWGMDLGSEHERYLTEQL-AKKPVIV 87
+ AAP + ++ A++ E +G+V W MD ++ + + L A K I
Sbjct 207 DTCAAPGMKTSHAAAIM-----ENQGKV-------WAMDRAADRVATMKQLLDASKVAIA 254
Query 88 HDYPKDIKAFYMKQNEDGKTVRAM-------DVLVPKIGELIGGSQREEDLEKLDRMIKE 140
+ D + + K A+ +V ++ E+ GG+ +E LEKL +
Sbjct 255 SSFCGDFLKTDVTDKKFSKVKFAIVDPPCSGSGIVKRMDEITGGNAEKERLEKLKNLQVH 314
Query 141 KGLNLDSYWW 150
K L+L W
Sbjct 315 KNLDLLKMCW 324
Lambda K H
0.318 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 3371754244
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40