bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0086_orf1
Length=329
Score E
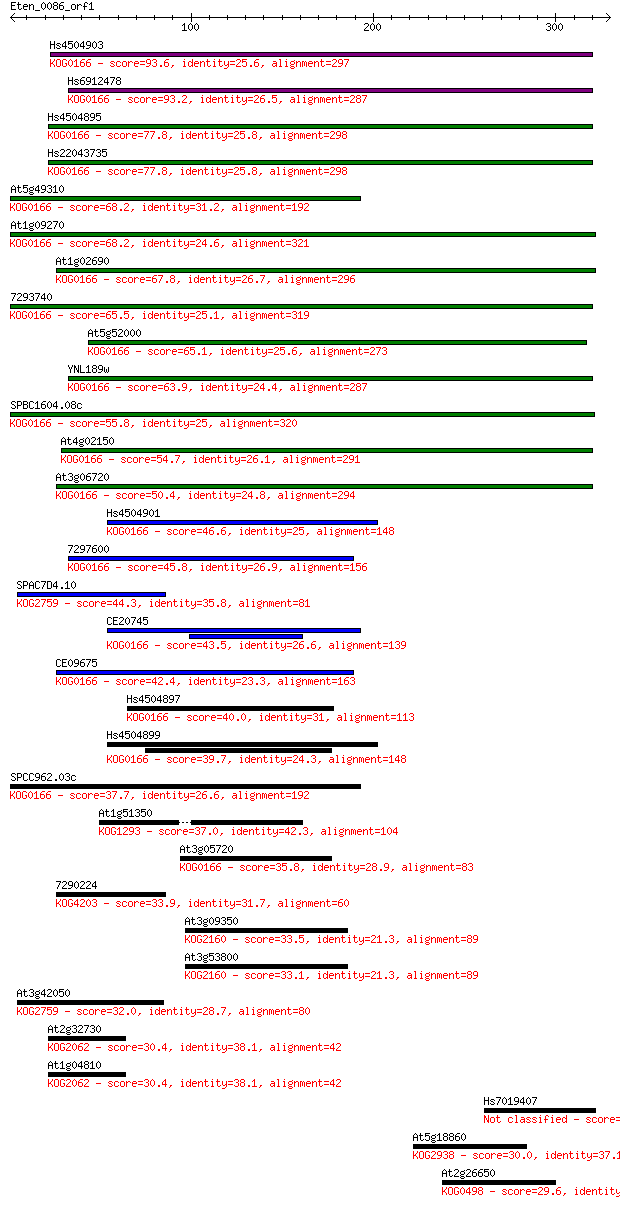
Sequences producing significant alignments: (Bits) Value
Hs4504903 93.6 6e-19
Hs6912478 93.2 7e-19
Hs4504895 77.8 3e-14
Hs22043735 77.8 3e-14
At5g49310 68.2 2e-11
At1g09270 68.2 3e-11
At1g02690 67.8 3e-11
7293740 65.5 2e-10
At5g52000 65.1 2e-10
YNL189w 63.9 4e-10
SPBC1604.08c 55.8 1e-07
At4g02150 54.7 3e-07
At3g06720 50.4 5e-06
Hs4504901 46.6 7e-05
7297600 45.8 1e-04
SPAC7D4.10 44.3 3e-04
CE20745 43.5 6e-04
CE09675 42.4 0.001
Hs4504897 40.0 0.007
Hs4504899 39.7 0.008
SPCC962.03c 37.7 0.031
At1g51350 37.0 0.058
At3g05720 35.8 0.13
7290224 33.9 0.55
At3g09350 33.5 0.71
At3g53800 33.1 0.76
At3g42050 32.0 1.7
At2g32730 30.4 5.0
At1g04810 30.4 5.1
Hs7019407 30.4 5.5
At5g18860 30.0 6.9
At2g26650 29.6 8.1
> Hs4504903
Length=536
Score = 93.6 bits (231), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 76/298 (25%), Positives = 131/298 (43%), Gaps = 51/298 (17%)
Query 23 LQFLVRVMSESTDPTSLSLACNACVTLCDSP-EGVSAVLEAMAMPHIMKLLCHSDETVVF 81
L L R++ S+DP L+ C A L D P + + AV+++ +++LL H+D VV
Sbjct 254 LNVLSRLLF-SSDPDVLADVCWALSYLSDGPNDKIQAVIDSGVCRRLVELLMHNDYKVVS 312
Query 82 DALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETP 141
AL I G Q ++ + ++ +L+ P + R AC T+ +
Sbjct 313 PALRAVGNIV-TGDDIQTQVILNCSALPCLLHLLSSPKES---IRKEACWTVSNITAGNR 368
Query 142 AQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVPSF 201
AQ+QA+ID FP+L+EI Q ++ TR EAA+A+ S +P+Q+ +V
Sbjct 369 AQIQAVIDANIFPVLIEILQ-KAEFRTRKEAAWAITNATSGGTPEQIRYLVALG------ 421
Query 202 MERGRGSSHVYFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLKLCKAILR 261
C+ + D+L ++ K+ + L
Sbjct 422 -----------------------------CIKPLCDLLTVMDS---------KIVQVALN 443
Query 262 GLENILEVGVQEGKSLDLPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSILQYF 319
GLENIL +G QE K + NPY L + G K+ ++ + + KA +++++
Sbjct 444 GLENILRLGEQESKQNGIGINPYCALIEEAYGLDKIEFLQSHENQEIYQKAFDLIEHY 501
> Hs6912478
Length=536
Score = 93.2 bits (230), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 76/288 (26%), Positives = 126/288 (43%), Gaps = 50/288 (17%)
Query 33 STDPTSLSLACNACVTLCDSP-EGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIA 91
S+D L+ AC A L D P E + AV+++ +++LL H+D V AL I
Sbjct 263 SSDSDLLADACWALSYLSDGPNEKIQAVIDSGVCRRLVELLMHNDYKVASPALRAVGNIV 322
Query 92 FIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQ 151
G Q ++ + ++ +L+ P + R AC TI + AQ+QA+ID
Sbjct 323 -TGDDIQTQVILNCSALPCLLHLLSSPKES---IRKEACWTISNITAGNRAQIQAVIDAN 378
Query 152 AFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVPSFMERGRGSSHV 211
FP+L+EI Q ++ TR EAA+A+ S +P+Q+ +V
Sbjct 379 IFPVLIEILQ-KAEFRTRKEAAWAITNATSGGTPEQIRYLV------------------- 418
Query 212 YFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLKLCKAILRGLENILEVGV 271
S C+ + D+L ++ K+ + L GLENIL +G
Sbjct 419 ----------------SLGCIKPLCDLLTVMDS---------KIVQVALNGLENILRLGE 453
Query 272 QEGKSLDLPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSILQYF 319
QEGK NPY L + G K+ ++ + + KA +++++
Sbjct 454 QEGKRSGSGVNPYCGLIEEAYGLDKIEFLQSHENQEIYQKAFDLIEHY 501
> Hs4504895
Length=538
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 77/299 (25%), Positives = 129/299 (43%), Gaps = 54/299 (18%)
Query 22 VLQFLVRVMSESTDPTSLSLACNACVTLCDSP-EGVSAVLEAMAMPHIMKLLCHSDETVV 80
VL +L+ V +D L+ AC A L D P + + AV++A +++LL H+D VV
Sbjct 258 VLSWLLFV----SDTDVLADACWALSYLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVV 313
Query 81 FDALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCET 140
AL I G Q ++ + +++ +L+ P + + AC TI +
Sbjct 314 SPALRAVGNIV-TGDDIQTQVILNCSALQSLLHLLSSPKES---IKKEACWTISNITAGN 369
Query 141 PAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVPS 200
AQ+Q +ID FP L+ I Q ++ TR EAA+A+ S S +Q+ +V
Sbjct 370 RAQIQTVIDANIFPALISILQT-AEFRTRKEAAWAITNATSGGSAEQIKYLV-------- 420
Query 201 FMERGRGSSHVYFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLKLCKAIL 260
E G C+ + D+L ++ K+ + L
Sbjct 421 --ELG-------------------------CIKPLCDLLTVMDS---------KIVQVAL 444
Query 261 RGLENILEVGVQEGKSLDLPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSILQYF 319
GLENIL +G QE K NPY L + G K+ ++ + + KA +++++
Sbjct 445 NGLENILRLGEQEAKRNGTGINPYCALIEEAYGLDKIEFLQSHENQEIYQKAFDLIEHY 503
> Hs22043735
Length=538
Score = 77.8 bits (190), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 77/299 (25%), Positives = 129/299 (43%), Gaps = 54/299 (18%)
Query 22 VLQFLVRVMSESTDPTSLSLACNACVTLCDSP-EGVSAVLEAMAMPHIMKLLCHSDETVV 80
VL +L+ V +D L+ AC A L D P + + AV++A +++LL H+D VV
Sbjct 258 VLSWLLFV----SDTDVLADACWALSYLSDGPNDKIQAVIDAGVCRRLVELLMHNDYKVV 313
Query 81 FDALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCET 140
AL I G Q ++ + +++ +L+ P + + AC TI +
Sbjct 314 SPALRAVGNIV-TGDDIQTQVILNCSALQSLLHLLSSPKES---IKKEACWTISNITAGN 369
Query 141 PAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVPS 200
AQ+Q +ID FP L+ I Q ++ TR EAA+A+ S S +Q+ +V
Sbjct 370 RAQIQTVIDANIFPALISILQT-AEFRTRKEAAWAITNATSGGSAEQIKYLV-------- 420
Query 201 FMERGRGSSHVYFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLKLCKAIL 260
E G C+ + D+L ++ K+ + L
Sbjct 421 --ELG-------------------------CIKPLCDLLTVMDS---------KIVQVAL 444
Query 261 RGLENILEVGVQEGKSLDLPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSILQYF 319
GLENIL +G QE K NPY L + G K+ ++ + + KA +++++
Sbjct 445 NGLENILRLGEQEAKRNGTGINPYCALIEEAYGLDKIEFLQSHENQEIYQKAFDLIEHY 503
> At5g49310
Length=519
Score = 68.2 bits (165), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 60/197 (30%), Positives = 94/197 (47%), Gaps = 11/197 (5%)
Query 1 RNSRWTLNT----KRTCLFADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCD-SPEG 55
RN+ WTL+ K + F D+ VL L R++ S D L AC A L D S E
Sbjct 217 RNATWTLSNFFRGKPSPPF-DLVKHVLPVLKRLVY-SDDEQVLIDACWALSNLSDASNEN 274
Query 56 VSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVL 115
+ +V+EA +P +++LL H+ V+ AL I G +QQ VI G + + +L
Sbjct 275 IQSVIEAGVVPRLVELLQHASPVVLVPALRCIGNIVS-GNSQQTHCVINCGVLPVLADLL 333
Query 116 TDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYA 175
T + R AC TI + Q+Q++ID P L+ + Q +++ + EA +A
Sbjct 334 TQNHMRG--IRREACWTISNITAGLEEQIQSVIDANLIPSLVNLAQ-HAEFDIKKEAIWA 390
Query 176 VCACLSRASPQQVGVIV 192
+ SP Q+ +V
Sbjct 391 ISNASVGGSPNQIKYLV 407
> At1g09270
Length=538
Score = 68.2 bits (165), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 79/327 (24%), Positives = 136/327 (41%), Gaps = 56/327 (17%)
Query 1 RNSRWTLNT----KRTCLFADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCDSP-EG 55
RN+ WTL+ K F +K A+ ++R + D L+ AC A L D P +
Sbjct 226 RNATWTLSNFCRGKPPTPFEQVKPALP--ILRQLIYLNDEEVLTDACWALSYLSDGPNDK 283
Query 56 VSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVL 115
+ AV+EA P +++LL H TV+ AL I G Q +I G + + +L
Sbjct 284 IQAVIEAGVCPRLVELLGHQSPTVLIPALRTVGNIV-TGDDSQTQFIIESGVLPHLYNLL 342
Query 116 TDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYA 175
T S + AC TI + Q++A++ L+ + Q + +++ + EAA+A
Sbjct 343 TQNHKKS--IKKEACWTISNITAGNKLQIEAVVGAGIILPLVHLLQ-NAEFDIKKEAAWA 399
Query 176 VCACLSRASPQQVGVIVCCTTRVPSFMERGRGSSHVYFAQELDEPCVLSLTGSNPCLHLI 235
+ S S +Q+ +V + C+ +
Sbjct 400 ISNATSGGSHEQIQYLV-----------------------------------TQGCIKPL 424
Query 236 ADMLELVSQSDPSNSGSLKLCKAILRGLENILEVGVQEGK-SLDLPENPYARLFQAVQGD 294
D+L DP ++ L GLENIL+VG + + L+ N YA++ + G
Sbjct 425 CDLLIC---PDP------RIVTVCLEGLENILKVGEADKEMGLNSGVNLYAQIIEESDGL 475
Query 295 IKLARMRYFPDYNVATKAHSILQYFTA 321
K+ ++ + + KA IL+ + A
Sbjct 476 DKVENLQSHDNNEIYEKAVKILERYWA 502
> At1g02690
Length=538
Score = 67.8 bits (164), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 79/309 (25%), Positives = 137/309 (44%), Gaps = 28/309 (9%)
Query 26 LVRVMSESTDPTSLSLACNACVTLCD----SPEGVSAVLEAMAMPHIMKLLCHSDETVVF 81
++ ++++ + + LS+ NA TL + P+ +A A+P + +LL +DE V+
Sbjct 206 MMSLLAQFHEHSKLSMLRNATWTLSNFCRGKPQPAFEQTKA-ALPALERLLHSTDEEVLT 264
Query 82 DALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETP 141
DA S A GT ++I VI G + +V++L P SP A TIG +
Sbjct 265 DA-SWALSYLSDGTNEKIQTVIDAGVIPRLVQLLAHP---SPSVLIPALRTIGNIVTGDD 320
Query 142 AQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQ-----QVGVI--VCC 194
Q QA+I +QA P LL + + + + EA + + + + Q Q G+I +
Sbjct 321 IQTQAVISSQALPGLLNLLKNTYKKSIKKEACWTISNITAGNTSQIQEVFQAGIIRPLIN 380
Query 195 TTRVPSFMERGRGSSHVYFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLK 254
+ F + + A + S C+ + D+L DP +
Sbjct 381 LLEIGEFEIKKEAVWAISNATSGGNHDQIKFLVSQGCIRPLCDLLPC---PDP------R 431
Query 255 LCKAILRGLENILEVGVQEGKSLDLP--ENPYARLFQAVQGDIKLARMRYFPDYNVATKA 312
+ L GLENIL+VG E K+L +N YA++ + G K+ ++ + + KA
Sbjct 432 VVTVTLEGLENILKVGEAE-KNLGNTGNDNLYAQMIEDADGLDKIENLQSHDNNEIYEKA 490
Query 313 HSILQYFTA 321
IL+ + A
Sbjct 491 VKILESYWA 499
> 7293740
Length=543
Score = 65.5 bits (158), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 80/326 (24%), Positives = 131/326 (40%), Gaps = 62/326 (19%)
Query 1 RNSRWTLNT-----KRTCLFADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCDSP-E 54
RN+ WTL+ FA + L L R++ + TD LS C A L D P +
Sbjct 241 RNAVWTLSNLCRGKSPPADFAKISHG-LPILARLL-KYTDADVLSDTCWAIGYLSDGPND 298
Query 55 GVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMVKV 114
+ AV++A +++LL H + V AL I G QQ ++G ++ + +
Sbjct 299 KIQAVIDAGVCRRLVELLLHPQQNVSTAALRAVGNIV-TGDDQQTQVILGYNALTCISHL 357
Query 115 LTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAY 174
L A + +C TI + Q+QA+I+ FP L+ I Q ++ TR EAA+
Sbjct 358 LHSTA---ETIKKESCWTISNIAAGNREQIQALINANIFPQLMVIMQT-AEFKTRKEAAW 413
Query 175 AVCACLSRASPQQVGVIVCCTTRVPSFMERGRGSSHVYFAQELDEPCVLSLTGSNPCLHL 234
A+ S + +Q+ +V C+
Sbjct 414 AITNATSSGTHEQIHYLVQVG-----------------------------------CVPP 438
Query 235 IADMLELVSQSDPSNSGSLKLCKAILRGLENILEVGVQEGKSLDLPENPYARLFQAVQGD 294
+ D L +V SD + + L LENIL+ G+ NPYA + G
Sbjct 439 MCDFLTVVD-SD--------IVQVALNALENILKA----GEKFQTRPNPYAITIEECGGL 485
Query 295 IKLARMRYFPDYNVATKAHSIL-QYF 319
K+ ++ + ++ K+ I+ QYF
Sbjct 486 DKIEYLQAHENRDIYHKSFYIIEQYF 511
> At5g52000
Length=441
Score = 65.1 bits (157), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 70/278 (25%), Positives = 120/278 (43%), Gaps = 55/278 (19%)
Query 44 NACVTLCD----SPEGVSAVLEAMAMPHIMKLLCHSDETVVFDAL-SIAARIAFIGTTQQ 98
NAC+ LC S +G+ +V+EA +P ++++L V+ AL +I A A G QQ
Sbjct 189 NACMALCHLSEGSEDGIQSVIEAGFVPKLVQILQLPSPVVLVPALLTIGAMTA--GNHQQ 246
Query 99 IDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLE 158
VI G + + +LT + AC I + T Q+Q++ID P+L+
Sbjct 247 TQCVINSGALPIISNMLTRNH--ENKIKKCACWVISNITAGTKEQIQSVIDANLIPILVN 304
Query 159 IFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVPSFMERGRGSSHVYFAQELD 218
+ Q D D+ + EA +A+ S Q+ Y A++
Sbjct 305 LAQ-DTDFYMKKEAVWAISNMALNGSHDQIK----------------------YMAEQ-- 339
Query 219 EPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLKLCKAILRGLENILEVGVQEGKSLD 278
C+ + D+L + ++ C L GLEN+L+ G E S D
Sbjct 340 -----------SCIKQLCDILVYSDER-----TTILKC---LDGLENMLKAGEAEKNSED 380
Query 279 LPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSIL 316
+ NPY L + +G K+++++ + ++ KA+ IL
Sbjct 381 V--NPYCLLIEDAEGLEKISKLQMNKNDDIYEKAYKIL 416
> YNL189w
Length=542
Score = 63.9 bits (154), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 70/291 (24%), Positives = 116/291 (39%), Gaps = 53/291 (18%)
Query 33 STDPTSLSLACNACVTLCDSP-EGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIA 91
S D +L AC A L D P E + AV++ +++LL H V AL I
Sbjct 268 SMDTETLVDACWAISYLSDGPQEAIQAVIDVRIPKRLVELLSHESTLVQTPALRAVGNIV 327
Query 92 FIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQ 151
G Q VI G + + +L+ P + AC TI + Q+QA+ID
Sbjct 328 -TGNDLQTQVVINAGVLPALRLLLSSP---KENIKKEACWTISNITAGNTEQIQAVIDAN 383
Query 152 AFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVPSFMERGRGSSHV 211
P L+++ +V +Y T+ EA +A+ S
Sbjct 384 LIPPLVKLLEV-AEYKTKKEACWAISNASSGG---------------------------- 414
Query 212 YFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSLKLCKAILRGLENILEVGV 271
L P ++ S C+ + D+LE+ ++ + L LENIL++G
Sbjct 415 -----LQRPDIIRYLVSQGCIKPLCDLLEIADN---------RIIEVTLDALENILKMGE 460
Query 272 --QEGKSLDLPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSILQ-YF 319
+E + L++ EN A + G K+ + + + KA+ I++ YF
Sbjct 461 ADKEARGLNINEN--ADFIEKAGGMEKIFNCQQNENDKIYEKAYKIIETYF 509
> SPBC1604.08c
Length=539
Score = 55.8 bits (133), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 80/332 (24%), Positives = 133/332 (40%), Gaps = 66/332 (19%)
Query 1 RNSRWTLNTKRTCL-------FADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCDSP 53
RN+ WTL+ C ++ + VAV L +++ S D + AC A L D P
Sbjct 224 RNATWTLSN--LCRGKNPPPNWSTISVAV-PILAKLLY-SEDVEIIVDACWAISYLSDGP 279
Query 54 -EGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMV 112
E + A+L+ P +++LL + AL I GT Q +I G ++
Sbjct 280 NEKIGAILDVGCAPRLVELLSSPSVNIQTPALRSVGNIV-TGTDAQTQIIIDCGALNAFP 338
Query 113 KVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEA 172
+L+ R AC TI + Q+QAII++ P L+ + DY T+ EA
Sbjct 339 SLLSH---QKENIRKEACWTISNITAGNTQQIQAIIESNLIPPLVHLLSY-ADYKTKKEA 394
Query 173 AYAVCACLSRA--SPQQVGVIVCCTTRVPSFMERGRGSSHVYFAQELDEPCVLSLTGSNP 230
+A+ S P Q+ +V S
Sbjct 395 CWAISNATSGGLGQPDQIRYLV-----------------------------------SQG 419
Query 231 CLHLIADMLELVSQSDPSNSGSLKLCKAILRGLENILEVGVQEGKSLDLPE-NPYARLFQ 289
+ + DML N K+ + L +ENIL+VG + +++DL N YA +
Sbjct 420 VIKPLCDML---------NGSDNKIIQVALDAIENILKVGEMD-RTMDLENINQYAVYVE 469
Query 290 AVQGDIKLARMRYFPDYNVATKAHSILQ-YFT 320
G + ++ + ++ KA+SI++ YF+
Sbjct 470 EAGGMDMIHDLQSSGNNDIYLKAYSIIEKYFS 501
> At4g02150
Length=531
Score = 54.7 bits (130), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 76/308 (24%), Positives = 130/308 (42%), Gaps = 36/308 (11%)
Query 29 VMSESTDPTSLSLACNACVTLCDSPEGVSA-VLEAM--AMPHIMKLLCHSDETVVFDALS 85
++S+ + T LS+ NA TL + G E A+P + +L+ DE V+ DA
Sbjct 208 LLSQFNENTKLSMLRNATWTLSNFCRGKPPPAFEQTQPALPVLERLVQSMDEEVLTDACW 267
Query 86 IAARIAFIGTTQQIDQVIGMGTVSTMVKVL--TDPAVASPMARARACNTIGYLGCETPAQ 143
+ ++ + +I VI G V ++++L + P+V P R TIG + Q
Sbjct 268 ALSYLS-DNSNDKIQAVIEAGVVPRLIQLLGHSSPSVLIPALR-----TIGNIVTGDDLQ 321
Query 144 VQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQ----------QVGVIVC 193
Q ++D QA P LL + + + + + EA + + + + Q Q V V
Sbjct 322 TQMVLDQQALPCLLNLLKNNYKKSIKKEACWTISNITAGNADQIQAVIDAGIIQSLVWVL 381
Query 194 CTTRVPSFMERGRGSSHVYFAQELDEPCVLSLTGSNPCLHLIADMLELVSQSDPSNSGSL 253
+ E G S+ D+ + S C+ + D+L L
Sbjct 382 QSAEFEVKKEAAWGISNATSGGTHDQ---IKFMVSQGCIKPLCDLL---------TCPDL 429
Query 254 KLCKAILRGLENILEVGVQEGKSLDL--PENPYARLFQAVQGDIKLARMRYFPDYNVATK 311
K+ L LENIL VG E K+L +N YA++ +G K+ ++ + ++ K
Sbjct 430 KVVTVCLEALENILVVGEAE-KNLGHTGEDNLYAQMIDEAEGLEKIENLQSHDNNDIYDK 488
Query 312 AHSILQYF 319
A IL+ F
Sbjct 489 AVKILETF 496
> At3g06720
Length=532
Score = 50.4 bits (119), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 73/319 (22%), Positives = 129/319 (40%), Gaps = 52/319 (16%)
Query 26 LVRVMSESTDPTSLSLACNACVTLCDSPEGVSAVLEAMAMPHI--MKLLCHSDETVVFDA 83
L+ ++++ + LS+ NA TL + G P + ++ L HSD+ V
Sbjct 202 LLPLLNQLNEHAKLSMLRNATWTLSNFCRGKPQPHFDQVKPALPALERLIHSDDEEVLTD 261
Query 84 LSIAARIAFIGTTQQIDQVIGMGTVSTMVKVL--TDPAVASPMARARACNTIGYLGCETP 141
A GT +I VI G V +V++L P+V P R T+G +
Sbjct 262 ACWALSYLSDGTNDKIQTVIQAGVVPKLVELLLHHSPSVLIPALR-----TVGNIVTGDD 316
Query 142 AQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQVGVIVCCTTRVP-- 199
Q Q +I++ A P L + + + + EA + + + ++ + Q+ +V P
Sbjct 317 IQTQCVINSGALPCLANLLTQNHKKSIKKEACWTI-SNITAGNKDQIQTVVEANLISPLV 375
Query 200 SFMERGR----------------GSSHVYFAQELDEPCVLSLTGSNPCLHLIADMLELVS 243
S ++ G SH +++ C+ L C +L+
Sbjct 376 SLLQNAEFDIKKEAAWAISNATSGGSHDQIKYLVEQGCIKPL-----C--------DLLV 422
Query 244 QSDPSNSGSLKLCKAILRGLENILEVGVQE---GKSLDLPENPYARLFQAVQGDIKLARM 300
DP ++ L GLENIL+VG E G + D+ N YA+L +G K+ +
Sbjct 423 CPDP------RIITVCLEGLENILKVGEAEKNLGHTGDM--NYYAQLIDDAEGLEKIENL 474
Query 301 RYFPDYNVATKAHSILQYF 319
+ + + KA IL+ +
Sbjct 475 QSHDNNEIYEKAVKILETY 493
> Hs4504901
Length=521
Score = 46.6 bits (109), Expect = 7e-05, Method: Compositional matrix adjust.
Identities = 37/148 (25%), Positives = 63/148 (42%), Gaps = 6/148 (4%)
Query 54 EGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMVK 113
E + V+++ +PH++ LL H + V AL I GT +Q V+ +S
Sbjct 275 EQIQMVIDSGIVPHLVPLLSHQEVKVQTAALRAVGNIV-TGTDEQTQVVLNCDALSHFPA 333
Query 114 VLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAA 173
+LT P A + + QVQA+ID P+++ + D+ T+ EAA
Sbjct 334 LLTHP---KEKINKEAVWFLSNITAGNQQQVQAVIDANLVPMIIHLLD-KGDFGTQKEAA 389
Query 174 YAVCACLSRASPQQVGVIVCCTTRVPSF 201
+A+ QV ++ +P F
Sbjct 390 WAISNLTISGRKDQVAYLIQQNV-IPPF 416
> 7297600
Length=522
Score = 45.8 bits (107), Expect = 1e-04, Method: Compositional matrix adjust.
Identities = 42/157 (26%), Positives = 71/157 (45%), Gaps = 6/157 (3%)
Query 33 STDPTSLSLACNACVTLCDSPE-GVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIA 91
S D L+ AC A + D + AV+++ A+P ++KLL + +++ AL I
Sbjct 251 SQDIQVLADACWALSYVTDDDNTKIQAVVDSDAVPRLVKLLQMDEPSIIVPALRSVGNI- 309
Query 92 FIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQ 151
GT QQ D VI + L S + + A T+ + Q+QA+I
Sbjct 310 VTGTDQQTDVVI--ASGGLPRLGLLLQHNKSNIVKEAAW-TVSNITAGNQKQIQAVIQAG 366
Query 152 AFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQV 188
F L + + D+ + EAA+AV + +P+Q+
Sbjct 367 IFQQLRTVLE-KGDFKAQKEAAWAVTNTTTSGTPEQI 402
> SPAC7D4.10
Length=450
Score = 44.3 bits (103), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 29/82 (35%), Positives = 43/82 (52%), Gaps = 4/82 (4%)
Query 5 WTLNTKRTCLFADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCDS-PEGVSAVLEAM 63
W N KR + A+L+ L ++ + D TSL++AC+ S PEG S +++
Sbjct 355 WHQNAKR---LNEDNYALLKKLFHIVQYNEDNTSLAVACHDLGAYIRSYPEGRSLIIKYG 411
Query 64 AMPHIMKLLCHSDETVVFDALS 85
A IM L+ H D V F+ALS
Sbjct 412 AKQRIMDLMSHPDPEVRFEALS 433
> CE20745
Length=514
Score = 43.5 bits (101), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 37/139 (26%), Positives = 58/139 (41%), Gaps = 5/139 (3%)
Query 54 EGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMVK 113
E + V+EA + H++ LL H D V AL I GT +Q V+ G + M
Sbjct 268 EHIQMVIEAQVVTHLVPLLGHVDVKVQTAALRAVGNIV-TGTDEQTQLVLDSGVLRFMPG 326
Query 114 VLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAA 173
+L A A + + QVQ + D P+++ + D+ T+ EAA
Sbjct 327 LL---AHYKEKINKEAVWFVSNITAGNQQQVQDVFDAGIMPMIIHLLDRG-DFPTQKEAA 382
Query 174 YAVCACLSRASPQQVGVIV 192
+A+ P QV +V
Sbjct 383 WAISNVTISGRPNQVEQMV 401
Score = 29.6 bits (65), Expect = 8.5, Method: Compositional matrix adjust.
Identities = 17/62 (27%), Positives = 30/62 (48%), Gaps = 3/62 (4%)
Query 99 IDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLE 158
ID +IG G + +V+ L+ P + A + + T Q QA+++ A PL L+
Sbjct 101 IDDLIGSGILPVLVQCLSS---TDPNLQFEAAWALTNIASGTSEQTQAVVNAGAVPLFLQ 157
Query 159 IF 160
+
Sbjct 158 LL 159
> CE09675
Length=531
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 38/164 (23%), Positives = 75/164 (45%), Gaps = 7/164 (4%)
Query 26 LVRVMSESTDPTSLSLACNACVTLCDSP-EGVSAVLEAMAMPHIMKLLCHSDETVVFDAL 84
LV+++ + TD AC A L D P E + E+ +PH++ + E +V AL
Sbjct 266 LVKLV-QHTDRQVRQDACWAVSYLTDGPDEQIELARESGVLPHVVAFFKEA-ENLVAPAL 323
Query 85 SIAARIAFIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQV 144
+A G VI +G++ ++ ++ +S + C + + T Q+
Sbjct 324 RTLGNVA-TGNDSLTQAVIDLGSLDEILPLMEKTRSSSIVKEC--CWLVSNIIAGTQKQI 380
Query 145 QAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAVCACLSRASPQQV 188
QA++D P+L+ + + D+ + EA++A+ + +QV
Sbjct 381 QAVLDANLLPVLINVLK-SGDHKCQFEASWALSNLAQGGTNRQV 423
> Hs4504897
Length=529
Score = 40.0 bits (92), Expect = 0.007, Method: Compositional matrix adjust.
Identities = 35/120 (29%), Positives = 58/120 (48%), Gaps = 19/120 (15%)
Query 65 MPHIMKLLCHSDETVVFDALSIAARIAFI--GTTQQIDQVIGMGTVSTMVKVL--TDPAV 120
+P +++LL H D V+ D I+++ G ++I V+ G V +VK+L ++ +
Sbjct 253 LPTLVRLLHHDDPEVLADT---CWAISYLTDGPNERIGMVVKTGVVPQLVKLLGASELPI 309
Query 121 ASPMARARACNTIGYLGCETPAQVQAIIDTQA---FPLLLEIFQVDPDYNTRIEAAYAVC 177
+P RA IG + T Q Q +ID A FP LL +P N + EA + +
Sbjct 310 VTPALRA-----IGNIVTGTDEQTQVVIDAGALAVFPSLL----TNPKTNIQKEATWTMS 360
> Hs4504899
Length=521
Score = 39.7 bits (91), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 36/148 (24%), Positives = 61/148 (41%), Gaps = 6/148 (4%)
Query 54 EGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMVK 113
E + V+++ +P ++ LL H + V AL I GT +Q V+ +S
Sbjct 275 EQIQMVIDSGVVPFLVPLLSHQEVKVQTAALRAVGNIV-TGTDEQTQVVLNCDVLSHFPN 333
Query 114 VLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAA 173
+L+ P A + + QVQA+ID P+++ D+ T+ EAA
Sbjct 334 LLSHP---KEKINKEAVWFLSNITAGNQQQVQAVIDAGLIPMIIHQL-AKGDFGTQKEAA 389
Query 174 YAVCACLSRASPQQVGVIVCCTTRVPSF 201
+A+ QV +V +P F
Sbjct 390 WAISNLTISGRKDQVEYLVQQNV-IPPF 416
Score = 30.4 bits (67), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 28/103 (27%), Positives = 46/103 (44%), Gaps = 4/103 (3%)
Query 75 SDETVV-FDALSIAARIAFIGTTQQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTI 133
SD VV A+ A ++ ID +I G + +VK L +P + A +
Sbjct 82 SDNPVVQLSAVQAARKLLSSDQNPPIDDLIKSGILPILVKCLERDD--NPSLQFEAAWAL 139
Query 134 GYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEAAYAV 176
+ T AQ QA++ + A PL L + + P N +A +A+
Sbjct 140 TNIASGTSAQTQAVVQSNAVPLFLRLLR-SPHQNVCEQAVWAL 181
> SPCC962.03c
Length=542
Score = 37.7 bits (86), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 51/202 (25%), Positives = 83/202 (41%), Gaps = 19/202 (9%)
Query 1 RNSRWTL-------NTKRTCLFADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCD-S 52
RNS WTL N + + VL L+ + E L A A L D +
Sbjct 224 RNSTWTLSNMCRGKNPQPDWNSISQVIPVLSKLIYTLDEDV----LVDALWAISYLSDGA 279
Query 53 PEGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAFIGTTQQIDQVIGMGTVSTMV 112
E + A+++A +++LL H V AL I G Q +I G +S ++
Sbjct 280 NEKIQAIIDAGIPRRLVELLMHPSAQVQTPALRSVGNIV-TGDDVQTQVIINCGALSALL 338
Query 113 KVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLLLEIFQVDPDYNTRIEA 172
+L+ P R AC TI + +Q+Q +I+ P L+ + D+ + EA
Sbjct 339 SLLSSPRDG---VRKEACWTISNITAGNSSQIQYVIEANIIPPLIHLLTT-ADFKIQKEA 394
Query 173 AYAVCACLSRAS--PQQVGVIV 192
+A+ S + P Q+ +V
Sbjct 395 CWAISNATSGGARRPDQIRYLV 416
> At1g51350
Length=666
Score = 37.0 bits (84), Expect = 0.058, Method: Compositional matrix adjust.
Identities = 19/43 (44%), Positives = 28/43 (65%), Gaps = 1/43 (2%)
Query 50 CDSPEGVSAVLEAMAMPHIMKLLCHSDETVVFDALSIAARIAF 92
C GV AVL+A PH+++LL ++DE VV DA + + R+ F
Sbjct 95 CGFEAGVQAVLDAGVFPHLLRLLTNTDEKVV-DAGARSLRMIF 136
Score = 34.3 bits (77), Expect = 0.40, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 35/73 (47%), Gaps = 12/73 (16%)
Query 100 DQVIGMGTVS-TMVKVLTDPAVASPMARAR---ACNTI--------GYLGCETPAQVQAI 147
+Q+IG T + +K+ PA+AS +A A CN I G C A VQA+
Sbjct 45 NQIIGNRTKKLSFLKLGAIPAIASVLADADDSDECNNILVQSAAALGSFACGFEAGVQAV 104
Query 148 IDTQAFPLLLEIF 160
+D FP LL +
Sbjct 105 LDAGVFPHLLRLL 117
> At3g05720
Length=528
Score = 35.8 bits (81), Expect = 0.13, Method: Compositional matrix adjust.
Identities = 24/85 (28%), Positives = 40/85 (47%), Gaps = 7/85 (8%)
Query 94 GTTQQIDQVIGMGTVSTMV--KVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQ 151
G+ ++I VI + ++ + P+V +P R TIG + +Q Q IID Q
Sbjct 260 GSNEKIQAVIEANVCARLIGLSIHRSPSVITPALR-----TIGNIVTGNDSQTQHIIDLQ 314
Query 152 AFPLLLEIFQVDPDYNTRIEAAYAV 176
A P L+ + + + R EA + V
Sbjct 315 ALPCLVNLLRGSYNKTIRKEACWTV 339
> 7290224
Length=840
Score = 33.9 bits (76), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 19/60 (31%), Positives = 32/60 (53%), Gaps = 0/60 (0%)
Query 26 LVRVMSESTDPTSLSLACNACVTLCDSPEGVSAVLEAMAMPHIMKLLCHSDETVVFDALS 85
LVR +S S D S A L +G+ A+ ++ +P ++KLL E+V+F A++
Sbjct 203 LVRAISNSNDLESTKAAVGTLHNLSHHRQGLLAIFKSGGIPALVKLLSSPVESVLFYAIT 262
> At3g09350
Length=387
Score = 33.5 bits (75), Expect = 0.71, Method: Compositional matrix adjust.
Identities = 19/89 (21%), Positives = 40/89 (44%), Gaps = 0/89 (0%)
Query 97 QQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLL 156
+ ID + ++ +V +L+ + RA+A + + + P + +++T A L
Sbjct 111 ESIDMANDLHSIGGLVPLLSFLKNSHANIRAKAADVVSTIVQNNPRSQELVMETNALESL 170
Query 157 LEIFQVDPDYNTRIEAAYAVCACLSRASP 185
L F D D + R +A A+ + + P
Sbjct 171 LSNFTSDTDIHARTQALGAISSLIRHNKP 199
> At3g53800
Length=363
Score = 33.1 bits (74), Expect = 0.76, Method: Compositional matrix adjust.
Identities = 19/89 (21%), Positives = 40/89 (44%), Gaps = 0/89 (0%)
Query 97 QQIDQVIGMGTVSTMVKVLTDPAVASPMARARACNTIGYLGCETPAQVQAIIDTQAFPLL 156
+ ID + ++ +V +L+ ++ RA++ + + + P Q +++ F L
Sbjct 87 ESIDLANDLHSIGGLVPLLSYLKNSNAKIRAKSADVLTTVVQNNPRSQQLVMEANGFEPL 146
Query 157 LEIFQVDPDYNTRIEAAYAVCACLSRASP 185
L F DPD R +A A+ + + P
Sbjct 147 LTNFIADPDIRVRTKALGAISSLIRNNQP 175
> At3g42050
Length=441
Score = 32.0 bits (71), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 23/81 (28%), Positives = 38/81 (46%), Gaps = 4/81 (4%)
Query 5 WTLNTKRTCLFADMKVAVLQFLVRVMSESTDPTSLSLACNACVTLCD-SPEGVSAVLEAM 63
W N TC F + +L+ L+ ++ S+DP SL++AC G V +
Sbjct 346 WRENV--TC-FEENDFQILRVLLTILDTSSDPRSLAVACFDISQFIQYHAAGRVIVADLK 402
Query 64 AMPHIMKLLCHSDETVVFDAL 84
A +MKL+ H + V +A+
Sbjct 403 AKERVMKLINHENAEVTKNAI 423
> At2g32730
Length=1004
Score = 30.4 bits (67), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 2/42 (4%)
Query 22 VLQFLVRVMSESTDPTSLSLACNACVTLCDSPEGVSAVLEAM 63
VL LV+V + P LS+ C C+ D P+GV+++LE +
Sbjct 201 VLSLLVKVYQKLPSPDYLSI-CQ-CLMFLDEPQGVASILEKL 240
> At1g04810
Length=1001
Score = 30.4 bits (67), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 16/42 (38%), Positives = 25/42 (59%), Gaps = 2/42 (4%)
Query 22 VLQFLVRVMSESTDPTSLSLACNACVTLCDSPEGVSAVLEAM 63
VL+ LV V + P LS+ C C+ D P+GV+++LE +
Sbjct 201 VLRLLVNVYQKLASPDYLSI-CQ-CLMFLDEPQGVASILEKL 240
> Hs7019407
Length=478
Score = 30.4 bits (67), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 18/61 (29%), Positives = 28/61 (45%), Gaps = 9/61 (14%)
Query 261 RGLENILEVGVQEGKSLDLPENPYARLFQAVQGDIKLARMRYFPDYNVATKAHSILQYFT 320
+G EN L+ E +S D+P+NP A L+ M F +Y + S+ +YF
Sbjct 319 KGTENKLKDDDFEEESFDIPDNPPASLY---------TNMNVFENYEASKAYSSVDEYFW 369
Query 321 A 321
Sbjct 370 G 370
> At5g18860
Length=890
Score = 30.0 bits (66), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 23/64 (35%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query 222 VLSLTGSNPCLHLIADMLELVSQSDP--SNSGSLKLCKAILRGLENILEVGVQEGKSLDL 279
+L + G + + DML L +QSDP G K KAI RG L+ G + DL
Sbjct 560 LLHMMGRDDIPVGLGDMLAL-NQSDPIFPPVGGCKYVKAIPRGCGGFLDSDTLYGLARDL 618
Query 280 PENP 283
P +P
Sbjct 619 PRSP 622
> At2g26650
Length=857
Score = 29.6 bits (65), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 18/65 (27%), Positives = 37/65 (56%), Gaps = 3/65 (4%)
Query 238 MLELVSQSDPS---NSGSLKLCKAILRGLENILEVGVQEGKSLDLPENPYARLFQAVQGD 294
+L L +DP+ GS+ L +A++ G E +++V ++ G ++D + + A QG+
Sbjct 568 LLLLEYHADPNCRDAEGSVPLWEAMVEGHEKVVKVLLEHGSTIDAGDVGHFACTAAEQGN 627
Query 295 IKLAR 299
+KL +
Sbjct 628 LKLLK 632
Lambda K H
0.322 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 7742285320
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40