bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
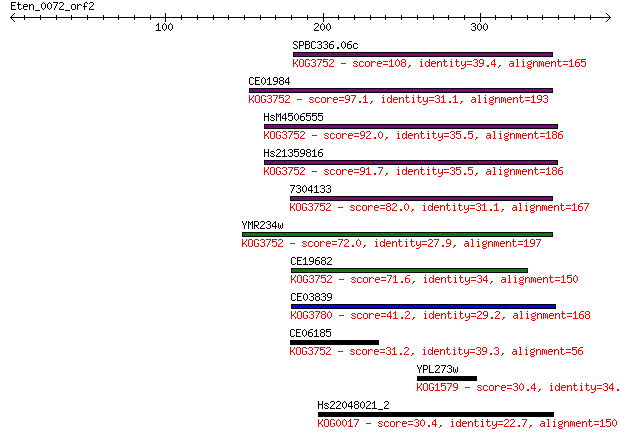
Query= Eten_0072_orf2
Length=382
Score E
Sequences producing significant alignments: (Bits) Value
SPBC336.06c 108 3e-23
CE01984 97.1 5e-20
HsM4506555 92.0 2e-18
Hs21359816 91.7 2e-18
7304133 82.0 2e-15
YMR234w 72.0 2e-12
CE19682 71.6 3e-12
CE03839 41.2 0.004
CE06185 31.2 3.7
YPL273w 30.4 6.8
Hs22048021_2 30.4 7.1
> SPBC336.06c
Length=264
Score = 108 bits (269), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 65/166 (39%), Positives = 92/166 (55%), Gaps = 30/166 (18%)
Query 181 IYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFVDY 240
+YADG+ NG+ A AG GVFFG DD R + +PL QT RAEL +I+ AL
Sbjct 126 VYADGSSLRNGKKGAVAGCGVFFGNDDPRNISVPLAGEEQTNNRAELQAIILAL------ 179
Query 241 DDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLI-QAI 299
+ SG L + SDS+Y++ L W +W +N +K+++ +PVKN DLI +A
Sbjct 180 ------ENTSG----DLTIRSDSNYSIKSLTTWLPKWKKNDFKTSNSQPVKNLDLINRAS 229
Query 300 LLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADKLAVQGA 345
L+ D+ +V EY++GHS YGN+ AD LA +GA
Sbjct 230 DLMSDR-------------NVSLEYVKGHSTDYGNQQADMLARRGA 262
> CE01984
Length=369
Score = 97.1 bits (240), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 60/193 (31%), Positives = 96/193 (49%), Gaps = 25/193 (12%)
Query 153 HRLLNCSPGASVHPQPETATPMKPDVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLF 212
H + + + E P + +Y DGAC SNG +A+AG GV++G D F
Sbjct 195 HEGTKFTEAKKMKTEEEVIDPEFANAPVVYTDGACSSNGTKNAKAGWGVYWGDDSEDNEF 254
Query 213 MPLVVGPQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGP 272
P V G T R EL ++ A+ + ++ K+ P ++V+ +DS+ V +
Sbjct 255 GP-VYGAPTNNRGELIAVQKAIEKAIE------KRLP------KVVIKTDSNLLVQSMNI 301
Query 273 WAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVY 332
W W R GWK+++G V N D++ I L K + V++ ++RGH+G+
Sbjct 302 WIHGWKRKGWKTSTGSEVLNQDVLMKIDNLRQKLK------------VKFLHVRGHAGID 349
Query 333 GNEMADKLAVQGA 345
GNE AD+LA +GA
Sbjct 350 GNEKADELARKGA 362
> HsM4506555
Length=286
Score = 92.0 bits (227), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 66/195 (33%), Positives = 97/195 (49%), Gaps = 40/195 (20%)
Query 163 SVHPQP----ETATPMKPDVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVG 218
SV P P +T + M D V +Y DG C SNGR RAGIGV++G PL VG
Sbjct 121 SVEPAPPVSRDTFSYMG-DFVVVYTDGCCSSNGRRRPRAGIGVYWGPGH------PLNVG 173
Query 219 -----PQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPW 273
QT QRAE+ + A+ +Q ++ +LV+ +DS + +N + W
Sbjct 174 IRLPGRQTNQRAEIHAACKAI------------EQAKTQNINKLVLYTDSMFTINGITNW 221
Query 274 AARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYG 333
W +NGWK+++GK V N + A+ L T +++ ++ GHSG G
Sbjct 222 VQGWKKNGWKTSAGKEVINKEDFVALERL------------TQGMDIQWMHVPGHSGFIG 269
Query 334 NEMADKLAVQGANSA 348
NE AD+LA +GA +
Sbjct 270 NEEADRLAREGAKQS 284
> Hs21359816
Length=286
Score = 91.7 bits (226), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 66/195 (33%), Positives = 97/195 (49%), Gaps = 40/195 (20%)
Query 163 SVHPQP----ETATPMKPDVVEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVG 218
SV P P +T + M D V +Y DG C SNGR RAGIGV++G PL VG
Sbjct 121 SVEPAPPVSRDTFSYMG-DFVVVYTDGCCSSNGRRRPRAGIGVYWGPGH------PLNVG 173
Query 219 -----PQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPW 273
QT QRAE+ + A+ +Q ++ +LV+ +DS + +N + W
Sbjct 174 IRLPGRQTNQRAEIHAACKAI------------EQAKTQNINKLVLYTDSMFTINGITNW 221
Query 274 AARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYG 333
W +NGWK+++GK V N + A+ L T +++ ++ GHSG G
Sbjct 222 VQGWKKNGWKTSAGKEVINKEDFVALERL------------TQGMDIQWMHVPGHSGFIG 269
Query 334 NEMADKLAVQGANSA 348
NE AD+LA +GA +
Sbjct 270 NEEADRLAREGAKQS 284
> 7304133
Length=333
Score = 82.0 bits (201), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 52/167 (31%), Positives = 79/167 (47%), Gaps = 23/167 (13%)
Query 179 VEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFV 238
V +Y DG+C NGR A AG GV+FG + + P V G T E+ + + A+ +
Sbjct 182 VIVYTDGSCIGNGRAGACAGYGVYFGKNHQLNAAKP-VEGRVTNNVGEIQAAIHAIKTAL 240
Query 239 DYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQA 298
D +L + +DS + +N + W A W + WK + +PVKN +
Sbjct 241 DLGIQ------------KLCISTDSQFLINSITLWVAGWKKRDWKLKNNQPVKNVVDFKE 288
Query 299 ILLLCDKRRKTAPSGATWRGSVEYEYIRGHSGVYGNEMADKLAVQGA 345
+ L + T V++ Y+ H G+ GNEMADKLA QG+
Sbjct 289 LDKLLQENNIT----------VKWNYVEAHKGIEGNEMADKLARQGS 325
> YMR234w
Length=348
Score = 72.0 bits (175), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 55/200 (27%), Positives = 90/200 (45%), Gaps = 15/200 (7%)
Query 149 AADHHRLLNCSPGASVHPQPETATPMKPDVVEIYADGACPSNGRVDARAGIGVFFGTDDR 208
+A ++L+N S + ++ M + +Y DG+ NG +RAG G +F
Sbjct 158 SAHDYKLMNISKESFESKYKLSSNTMYNKSMNVYCDGSSFGNGTSSSRAGYGAYFEGAPE 217
Query 209 RYLFMPLVVGPQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVRLVVISDSSYAVN 268
+ PL+ G QT RAE+ ++ AL + + + + + +V + +DS Y
Sbjct 218 ENISEPLLSGAQTNNRAEIEAVSEALKKIWE-------KLTNEKEKVNYQIKTDSEYVTK 270
Query 269 CLGPWAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATW---RGSVEYEYI 325
L R+ K G P N+DLI ++ K +K G + E++
Sbjct 271 LLND---RYMTYDNKKLEGLP--NSDLIVPLVQRFVKVKKYYELNKECFKNNGKFQIEWV 325
Query 326 RGHSGVYGNEMADKLAVQGA 345
+GH G GNEMAD LA +GA
Sbjct 326 KGHDGDPGNEMADFLAKKGA 345
> CE19682
Length=139
Score = 71.6 bits (174), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 51/150 (34%), Positives = 72/150 (48%), Gaps = 19/150 (12%)
Query 180 EIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFVD 239
+ Y DG+ NG+ +R G GV +D + + GPQT R EL +I A D
Sbjct 4 DFYTDGSALGNGQQGSRGGYGVHVPSDTSKNVSGSYPHGPQTNNRYELEAIKHATQMARD 63
Query 240 YDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQAI 299
D S VR + +DS A + + W W NG+K++SG+ VKN DLI+ I
Sbjct 64 VPD----------SHVR--IHTDSKNAKDSVTKWNDNWKSNGYKTSSGQDVKNQDLIRDI 111
Query 300 LLLCDKRRKTAPSGATWRGSVEYEYIRGHS 329
+ SG T VE+E++RGHS
Sbjct 112 DRNV---QALKHSGKT----VEFEHVRGHS 134
> CE03839
Length=404
Score = 41.2 bits (95), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 49/171 (28%), Positives = 69/171 (40%), Gaps = 12/171 (7%)
Query 180 EIYADGACPSNGRVDARAGIGVFFGTDDR--RYLFMPLVVGPQTYQRAELASILAALLRF 237
E+Y DG+ +NG+ AR G + F D Y FM VG QT EL +I A
Sbjct 7 EVYTDGSTVNNGKRGARGGWAIVFPFDRSLDEYDFMK--VGKQTNNVYELTAIYEATEVV 64
Query 238 VDYDDDSAKQQPSGRSEVRLVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQ 297
+D Q+ + + SS + C + NG ST G + +
Sbjct 65 RCWDSFKCNQKLEFVRLKNTITPTTSSILIPCTRRTVS---PNG--STDGTNMGGLRITP 119
Query 298 AILLLCDKRRKTAPSGATWRG-SVEYEYIRGHSGVYGNEMADKLAVQGANS 347
I L R PS A V+ EY++GH + N AD+LA + S
Sbjct 120 GI--LSKISRSFVPSTAISESLDVKIEYVKGHHTNFFNCEADRLAKKACRS 168
> CE06185
Length=648
Score = 31.2 bits (69), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 22/56 (39%), Positives = 30/56 (53%), Gaps = 3/56 (5%)
Query 179 VEIYADGACPSNGRVDARAGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAAL 234
V I+A C G+ DA A GV++G DD R LV QT RA + ++ +AL
Sbjct 166 VAIFA--ICEKEGKCDALAKYGVYWGQDDHRN-EAGLVEDGQTSLRAIMCAVRSAL 218
> YPL273w
Length=325
Score = 30.4 bits (67), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 13/38 (34%), Positives = 18/38 (47%), Gaps = 1/38 (2%)
Query 260 ISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQ 297
I + Y + C+GPW A CR +P +N D Q
Sbjct 116 IGEDKYLIGCIGPWGAHICREFTGDYGAEP-ENIDFYQ 152
> Hs22048021_2
Length=835
Score = 30.4 bits (67), Expect = 7.1, Method: Compositional matrix adjust.
Identities = 34/150 (22%), Positives = 59/150 (39%), Gaps = 24/150 (16%)
Query 197 AGIGVFFGTDDRRYLFMPLVVGPQTYQRAELASILAALLRFVDYDDDSAKQQPSGRSEVR 256
AG G++ + + + P T A LA++ L RF G+S +
Sbjct 262 AGFGLYVLSPTSPPVSLSFSCSPYTPTYAHLAAVACGLERF-------------GQSPLP 308
Query 257 LVVISDSSYAVNCLGPWAARWCRNGWKSTSGKPVKNADLIQAILLLCDKRRKTAPSGATW 316
+V ++ ++ + L W G+ S+ G P+ + L+ I+ L SG +
Sbjct 309 VVFLTHCNWIFSLLWELLPLWRARGFLSSDGAPLPHPSLLSYIISLT--------SGLS- 359
Query 317 RGSVEYEYIRGHSGVYGNEMADKLAVQGAN 346
S+ + Y + G D LA QGA
Sbjct 360 --SLPFIYRTSYRGSLFAVTVDTLAKQGAQ 387
Lambda K H
0.318 0.134 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 9541565792
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40