bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
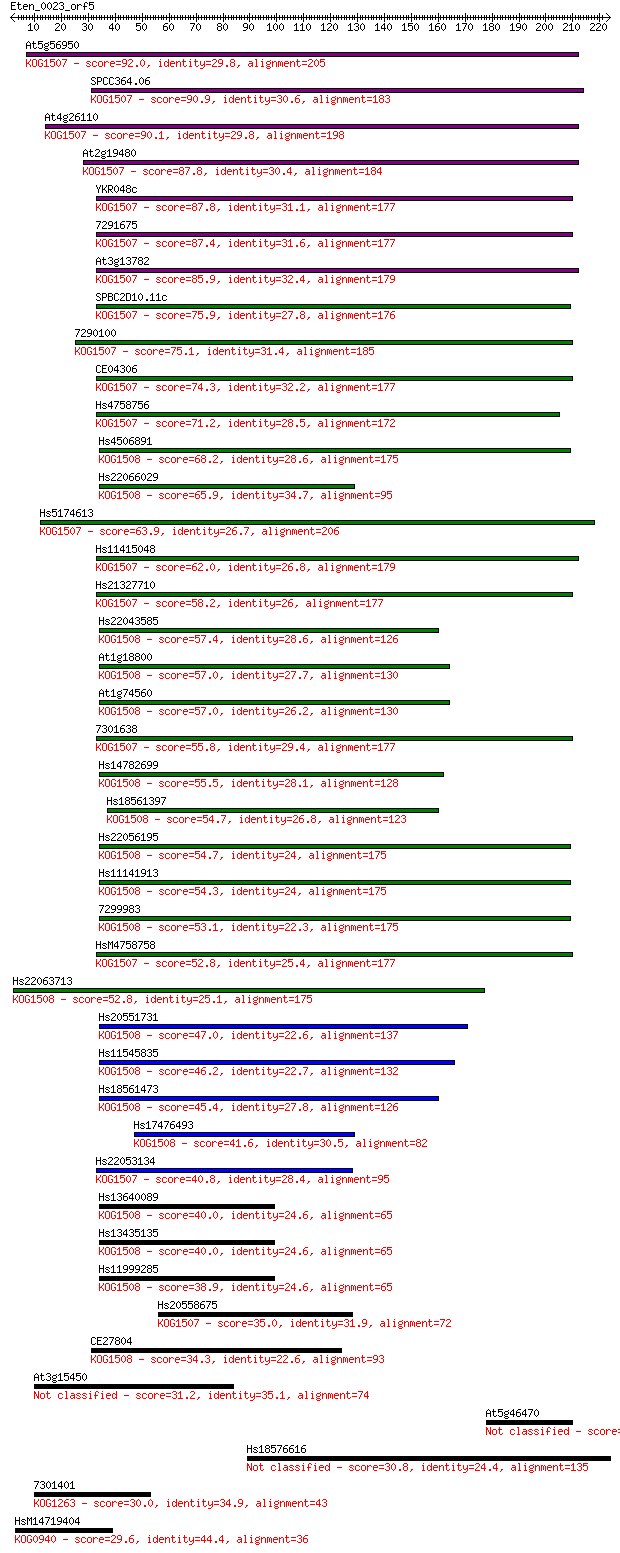
Query= Eten_0023_orf5
Length=223
Score E
Sequences producing significant alignments: (Bits) Value
At5g56950 92.0 9e-19
SPCC364.06 90.9 2e-18
At4g26110 90.1 3e-18
At2g19480 87.8 1e-17
YKR048c 87.8 1e-17
7291675 87.4 2e-17
At3g13782 85.9 7e-17
SPBC2D10.11c 75.9 6e-14
7290100 75.1 1e-13
CE04306 74.3 2e-13
Hs4758756 71.2 1e-12
Hs4506891 68.2 1e-11
Hs22066029 65.9 7e-11
Hs5174613 63.9 2e-10
Hs11415048 62.0 9e-10
Hs21327710 58.2 1e-08
Hs22043585 57.4 2e-08
At1g18800 57.0 3e-08
At1g74560 57.0 3e-08
7301638 55.8 7e-08
Hs14782699 55.5 8e-08
Hs18561397 54.7 1e-07
Hs22056195 54.7 1e-07
Hs11141913 54.3 2e-07
7299983 53.1 5e-07
HsM4758758 52.8 5e-07
Hs22063713 52.8 6e-07
Hs20551731 47.0 3e-05
Hs11545835 46.2 5e-05
Hs18561473 45.4 8e-05
Hs17476493 41.6 0.001
Hs22053134 40.8 0.002
Hs13640089 40.0 0.004
Hs13435135 40.0 0.004
Hs11999285 38.9 0.009
Hs20558675 35.0 0.11
CE27804 34.3 0.18
At3g15450 31.2 1.7
At5g46470 30.8 2.1
Hs18576616 30.8 2.5
7301401 30.0 4.0
HsM14719404 29.6 4.7
> At5g56950
Length=368
Score = 92.0 bits (227), Expect = 9e-19, Method: Compositional matrix adjust.
Identities = 61/205 (29%), Positives = 97/205 (47%), Gaps = 12/205 (5%)
Query 7 AAPENAAAAEAETAVTPSAGYSVATPGLPGFWLSVLEREAHTARSLSSGDRACLLYLEDV 66
APE+A + + + G+P FWL+ L+ + ++ D L+YL+D+
Sbjct 106 GAPEDAKMDQGDEK-------TAEEKGVPSFWLTALKNNDVISEEITERDEGALIYLKDI 158
Query 67 RADKRLRKEDGFSVSFVFAANPFFSNRVLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDL 126
+ K + + GF + F F NP+F N +L+K M + D+ ++ K IGTEI W G L
Sbjct 159 KWCK-IEEPKGFKLEFFFDQNPYFKNTLLTKAYHMIDE-DEPLLEKAIGTEIDWYPGKCL 216
Query 127 TKSVRISKQRNLRTREVRIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLS 186
T+ + K + I K E SFFNFF QVP + A L +
Sbjct 217 TQKILKKKPKKGAKNAKPITKTEDCE-SFFNFFNPPQVPDDDEDIDEE--RAEELQNLME 273
Query 187 TEYEMAVVIRDIIVPKAIGVYLGVA 211
+Y++ IR+ I+P A+ + G A
Sbjct 274 QDYDIGSTIREKIIPHAVSWFTGEA 298
> SPCC364.06
Length=393
Score = 90.9 bits (224), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 56/186 (30%), Positives = 93/186 (50%), Gaps = 9/186 (4%)
Query 31 TPGLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFF 90
T G+P FWL+ ++ + ++ D L +L D+R ++ GF + F FA NPFF
Sbjct 161 TKGIPEFWLTAMKNVLSLSEMITPEDEGALSHLVDIRIS--YMEKPGFKLEFEFAENPFF 218
Query 91 SNRVLSKEIC-MQKQGDDAVVA--KTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVK 147
+N++L+K M++ G V G ++ W E DLT KQRN T++ R+VK
Sbjct 219 TNKILTKTYYYMEESGPSNVFLYDHAEGDKVDWKENADLTVRTVTKKQRNKNTKQTRVVK 278
Query 148 NITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVY 207
+SFFNFF P + E+ L + L +Y++ ++ ++P+A+ +
Sbjct 279 VSVPRDSFFNFFN----PPTPPSEEDEESESPELDELLELDYQIGEDFKEKLIPRAVEWF 334
Query 208 LGVAGA 213
G A A
Sbjct 335 TGEALA 340
> At4g26110
Length=372
Score = 90.1 bits (222), Expect = 3e-18, Method: Compositional matrix adjust.
Identities = 59/198 (29%), Positives = 94/198 (47%), Gaps = 5/198 (2%)
Query 14 AAEAETAVTPSAGYSVATPGLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLR 73
A E +T V + G+P FWL+ L+ + ++ D L YL+D++ K +
Sbjct 107 APEDDTKVDQGEEKTAEEKGVPSFWLTALKNNDVISEEVTERDEGALKYLKDIKWCK-IE 165
Query 74 KEDGFSVSFVFAANPFFSNRVLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRIS 133
+ GF + F F NP+F N VL+K M + D+ ++ K +GTEI W G LT+ +
Sbjct 166 EPKGFKLEFFFDTNPYFKNTVLTKSYHMIDE-DEPLLEKAMGTEIDWYPGKCLTQKILKK 224
Query 134 KQRNLRTREVRIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAV 193
K + I K + SFFNFF +VP A L + +Y++
Sbjct 225 KPKKGSKNTKPITK-LEDCESFFNFFSPPEVPDEDEDIDEE--RAEDLQNLMEQDYDIGS 281
Query 194 VIRDIIVPKAIGVYLGVA 211
IR+ I+P+A+ + G A
Sbjct 282 TIREKIIPRAVSWFTGEA 299
> At2g19480
Length=379
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 56/184 (30%), Positives = 88/184 (47%), Gaps = 5/184 (2%)
Query 28 SVATPGLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAAN 87
S G+P FWL L+ TA ++ D L YL+D++ R+ + GF + F F N
Sbjct 120 SAEEKGVPDFWLIALKNNEITAEEITERDEGALKYLKDIKWS-RVEEPKGFKLEFFFDQN 178
Query 88 PFFSNRVLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVK 147
P+F N VL+K M + D+ ++ K +GTEI W G LT+ + K + I K
Sbjct 179 PYFKNTVLTKTYHMIDE-DEPILEKALGTEIEWYPGKCLTQKILKKKPKKGSKNTKPITK 237
Query 148 NITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVY 207
E SFFNFF QVP + + ++ +Y++ I++ I+ A+ +
Sbjct 238 TEDCE-SFFNFFSPPQVPDDDEDLDDDMADELQ--GQMEHDYDIGSTIKEKIISHAVSWF 294
Query 208 LGVA 211
G A
Sbjct 295 TGEA 298
> YKR048c
Length=417
Score = 87.8 bits (216), Expect = 1e-17, Method: Compositional matrix adjust.
Identities = 55/183 (30%), Positives = 94/183 (51%), Gaps = 10/183 (5%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVF--AANPFF 90
G+P FWL+ LE +++ D L YL+D+ + GF + F F +ANPFF
Sbjct 184 GIPSFWLTALENLPIVCDTITDRDAEVLEYLQDIGLEYLTDGRPGFKLLFRFDSSANPFF 243
Query 91 SNRVLSKEICMQKQ---GDDAVVAKTIGTEIHWNEG-MDLTKSVRISKQRNLRTREVRIV 146
+N +L K QK+ D + G EI W + ++T + + KQRN T++VR +
Sbjct 244 TNDILCKTYFYQKELGYSGDFIYDHAEGCEISWKDNAHNVTVDLEMRKQRNKTTKQVRTI 303
Query 147 KNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGV 206
+ IT SFFNFF+ ++ + + L +RL+ +Y + ++D ++P+A+
Sbjct 304 EKITPIESFFNFFDPPKIQNEDQDEEL----EEDLEERLALDYSIGEQLKDKLIPRAVDW 359
Query 207 YLG 209
+ G
Sbjct 360 FTG 362
> 7291675
Length=370
Score = 87.4 bits (215), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 56/188 (29%), Positives = 97/188 (51%), Gaps = 22/188 (11%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDG--FSVSFVFAANPFF 90
G+PGFWL+V A + + D + L D+ ++ ++G +++ F F N +F
Sbjct 145 GIPGFWLTVFRNTAIMSEMVQPHDEPAIRKLIDIS----IKYDNGHSYTLEFHFDKNEYF 200
Query 91 SNRVLSKEICMQKQGDD--------AVVAKTIGTEIHWNEGMDLT-KSVRISKQRNLRTR 141
SN VL+K+ ++ D + K G I+W + M+LT K++R KQ++
Sbjct 201 SNSVLTKQYVLKSTVDPNDPFAFEGPEIYKCTGCTINWEKKMNLTVKTIR-KKQKHKERG 259
Query 142 EVRIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVP 201
VR + +SFFNFF +VPS++ + DD ++ L+T++E+ +R I+P
Sbjct 260 AVRTIVKQVPTDSFFNFFSPPEVPSDQ--EEVDDDSQQI----LATDFEIGHFLRARIIP 313
Query 202 KAIGVYLG 209
KA+ Y G
Sbjct 314 KAVLYYTG 321
> At3g13782
Length=329
Score = 85.9 bits (211), Expect = 7e-17, Method: Compositional matrix adjust.
Identities = 58/184 (31%), Positives = 95/184 (51%), Gaps = 6/184 (3%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSN 92
G+P FWL ++ A ++ D A L YL+D+R+ + F + F+F +N +F N
Sbjct 127 GVPNFWLIAMKTNEMLANEITERDEAALKYLKDIRSCRVEDTSRNFKLEFLFDSNLYFKN 186
Query 93 RVLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSE 152
VLSK + + D V+ K IGT+I W G LT V + K+ ++V + +E
Sbjct 187 SVLSKTYHVNDE-DGPVLEKVIGTDIEWFPGKCLTHKVVVKKKTKKGPKKVNNIPMTKTE 245
Query 153 N--SFFNFFENTQVPSNRAIRAMGDDEARL---LFKRLSTEYEMAVVIRDIIVPKAIGVY 207
N SFFNFF+ ++P + D + + L + +Y++AV IRD ++P A+ +
Sbjct 246 NCESFFNFFKPPEIPEIDEVDDYDDFDTIMTEELQNLMDQDYDIAVTIRDKLIPHAVSWF 305
Query 208 LGVA 211
G A
Sbjct 306 TGEA 309
> SPBC2D10.11c
Length=379
Score = 75.9 bits (185), Expect = 6e-14, Method: Compositional matrix adjust.
Identities = 49/180 (27%), Positives = 84/180 (46%), Gaps = 9/180 (5%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSN 92
G+P FWL+ L ++ D L L D+R G+ + F F +N +F+N
Sbjct 165 GIPEFWLTCLHNVFLVGEMITPEDENVLRSLSDIRFTNLSGDVHGYKLEFEFDSNDYFTN 224
Query 93 RVLSKEICMQKQ---GDDAVVAKTIGTEIHW-NEGMDLTKSVRISKQRNLRTREVRIVKN 148
++L+K + + + G +I+W +LT V KQRN +T + R+V+
Sbjct 225 KILTKTYYYKDDLSPSGEFLYDHAEGDKINWIKPEKNLTVRVETKKQRNRKTNQTRLVRT 284
Query 149 ITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVYL 208
+SFFNFF Q+ + + + D + L +Y++ V +D I+P AI +L
Sbjct 285 TVPNDSFFNFFSPPQLDDDESDDGLDDKT-----ELLELDYQLGEVFKDQIIPLAIDCFL 339
> 7290100
Length=362
Score = 75.1 bits (183), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 58/195 (29%), Positives = 89/195 (45%), Gaps = 24/195 (12%)
Query 25 AGYSVATPGLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVF 84
A S T G+P FWL+V + + + D L L DVR +D + V F F
Sbjct 156 APVSPTTLGVPRFWLTVFQNVPLLSELVQDHDEPLLESLMDVRLAY---DQDSYMVIFQF 212
Query 85 AANPFF--SNRVLSKEICMQKQGDDAV--------VAKTIGTEIHWNEGMDLTKSVRISK 134
N F S+ +L+K +Q D + + G IHW +G +LT S+
Sbjct 213 RPNSFLHDSSLLLTKRYFLQHSADPEYPFLFEGPEIVRCEGCHIHWRDGSNLTLQTVESR 272
Query 135 QRNLRTREVRIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVV 194
+RN R R+ K + E SFF FF P ++ D++ +L+ L ++E+ +
Sbjct 273 RRN---RAHRVTKVMPRE-SFFRFF----APPQALDLSLADEKTKLI---LGNDFEVGFL 321
Query 195 IRDIIVPKAIGVYLG 209
+R IVPKA+ Y G
Sbjct 322 LRTQIVPKAVLFYTG 336
> CE04306
Length=316
Score = 74.3 bits (181), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 57/185 (30%), Positives = 84/185 (45%), Gaps = 14/185 (7%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSN 92
G+ FWL+ L A ++ D L YL DV + GF + F FA NP+F N
Sbjct 112 GIKDFWLTALRTHDLVAEAIEEHDVPILSYLTDVTTAAS-KDPAGFKIEFHFATNPYFKN 170
Query 93 RVLSKEICM--------QKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVR 144
+VL+K + Q D V + +G I W +G ++TK KQ+
Sbjct 171 QVLTKTYLLGFDPDAEAPLQFDGPHVIRAVGDTIEWEDGKNVTKKAVKKKQKKGANAGKF 230
Query 145 IVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAI 204
+ K + + +SFFNFFE P D++ + L +YEM IRD I+P+A+
Sbjct 231 LTKTVKA-DSFFNFFE----PPKSKDERNEDEDDEQAEEFLELDYEMGQAIRDTIIPRAV 285
Query 205 GVYLG 209
Y G
Sbjct 286 LFYTG 290
> Hs4758756
Length=391
Score = 71.2 bits (173), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 49/181 (27%), Positives = 89/181 (49%), Gaps = 16/181 (8%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRAD-KRLRKEDGFSVSFVFAANPFFS 91
G+P FWL+V + + + D L +L+D++ + F + F F N +F+
Sbjct 166 GIPEFWLTVFKNVDLLSDMVQEHDEPILKHLKDIKVKFSDAGQPMSFVLEFHFEPNEYFT 225
Query 92 NRVLSKEICMQKQGDDA--------VVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREV 143
N VL+K M+ + DD+ + G +I W +G ++T KQ++ V
Sbjct 226 NEVLTKTYRMRSEPDDSDPFSFDGPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKGRGTV 285
Query 144 RIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKA 203
R V S +SFFNFF +VP + + DD+A + L+ ++E+ +R+ I+P++
Sbjct 286 RTVTKTVSNDSFFNFFAPPEVPESGDL----DDDAEAI---LAADFEIGHFLRERIIPRS 338
Query 204 I 204
+
Sbjct 339 V 339
> Hs4506891
Length=277
Score = 68.2 bits (165), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 50/175 (28%), Positives = 74/175 (42%), Gaps = 31/175 (17%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+P FW++ + L D L YL V + + G+ + F F NP+F N+
Sbjct 78 IPNFWVTTFVNHPQVSALLGEEDEEALHYLTRVEVTEFEDIKSGYRIDFYFDENPYFENK 137
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
VLSKE + + GD + + TEI W G DLTK R S+ +N +R+ +
Sbjct 138 VLSKEFHLNESGDPS----SKSTEIKWKSGKDLTK--RSSQTQNKASRK----RQHEEPE 187
Query 154 SFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVYL 208
SFF +F + G D E+ VI+D I P + YL
Sbjct 188 SFFTWFTDH--------SDAGAD-------------ELGEVIKDDIWPNPLQYYL 221
> Hs22066029
Length=442
Score = 65.9 bits (159), Expect = 7e-11, Method: Compositional matrix adjust.
Identities = 33/95 (34%), Positives = 46/95 (48%), Gaps = 4/95 (4%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+P FW++ + L D L YL V + + G+ + F F NP+ N+
Sbjct 349 IPHFWITTFVNHPQVSALLGEEDEEALHYLTRVEVTEFEDIKSGYRIDFYFDQNPYLENK 408
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTK 128
VLSKE + K GD + + TEI W GMDLTK
Sbjct 409 VLSKEFHLNKSGDPS----SKSTEIKWKSGMDLTK 439
> Hs5174613
Length=375
Score = 63.9 bits (154), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 55/222 (24%), Positives = 95/222 (42%), Gaps = 26/222 (11%)
Query 12 AAAAEAETAVTPSAGYSVATP---GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRA 68
A +++ VT A + P G+P FW ++ + + D L +L+D++
Sbjct 134 AGDMKSKVVVTEKAAATAEEPDPKGIPEFWFTIFRNVDMLSELVQEYDEPILKHLQDIKV 193
Query 69 D-KRLRKEDGFSVSFVFAANPFFSNRVLSKEICMQKQGDDA--------VVAKTIGTEIH 119
+ F + F F N +F+N VL+K M+ + D A + G I
Sbjct 194 KFSDPGQPMSFVLEFHFEPNDYFTNSVLTKTYKMKSEPDKADPFSFEGPEIVDCDGCTID 253
Query 120 WNEGMDLTKSVRISKQRNLRTREVRIVKNITSENSFFNFFENTQVPSNRAIRAMGD---- 175
W +G ++T KQ++ VR + SFFNFF ++A GD
Sbjct 254 WKKGKNVTVKTIKKKQKHKGRGTVRTITKQVPNESFFNFF--------NPLKASGDGESL 305
Query 176 DEARLLFKRLSTEYEMAVVIRDIIVPKAIGVYLGVAGAESDS 217
DE L++++E+ R+ IVP+A+ + G A + D+
Sbjct 306 DEDSEF--TLASDFEIGHFFRERIVPRAVLYFTGEAIEDDDN 345
> Hs11415048
Length=460
Score = 62.0 bits (149), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 48/188 (25%), Positives = 80/188 (42%), Gaps = 23/188 (12%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKED-GFSVSFVFAANPFFS 91
G+P FWL+VL+ + D L L D++ E F++ F F N +F
Sbjct 239 GIPDFWLTVLKNVDTLTPLIKKYDEPILKLLTDIKVKLSDPGEPLSFTLEFHFKPNEYFK 298
Query 92 NRVLSKEICMQKQ--------GDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREV 143
N +L+K ++ + + + G EI WNEG ++T KQ++ +
Sbjct 299 NELLTKTYVLKSKLAYYDPHPYRGTAIEYSTGCEIDWNEGKNVTLKTIKKKQKHRIWGTI 358
Query 144 RIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKA 203
R V ++SFFNFF + SN R DD + + +R I+P++
Sbjct 359 RTVTEDFPKDSFFNFFSPHGITSNG--RDGNDD------------FLLGHNLRTYIIPRS 404
Query 204 IGVYLGVA 211
+ + G A
Sbjct 405 VLFFSGDA 412
> Hs21327710
Length=506
Score = 58.2 bits (139), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 46/186 (24%), Positives = 78/186 (41%), Gaps = 18/186 (9%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRAD-KRLRKEDGFSVSFVFAANPFFS 91
G+P +WL VL+ + D L +L DV + + ++ F F NP+F
Sbjct 310 GIPDYWLIVLKNVDKLGPMIQKYDEPILKFLSDVSLKFSKPGQPVSYTFEFHFLPNPYFR 369
Query 92 NRVLSKEICMQKQGDDA--------VVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREV 143
N VL K ++ + D + G +I W G D+T + ++ R T E+
Sbjct 370 NEVLVKTYIIKAKPDHNDPFFSWGWEIEDCKGCKIDWRRGKDVTVTT--TQSRTTATGEI 427
Query 144 RIVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKA 203
I + SFFNFF ++P + D L ++E+ ++ D ++ K+
Sbjct 428 EIQPRVVPNASFFNFFSPPEIPMIGKLEPRED-------AILDEDFEIGQILHDNVILKS 480
Query 204 IGVYLG 209
I Y G
Sbjct 481 IYYYTG 486
> Hs22043585
Length=250
Score = 57.4 bits (137), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 36/126 (28%), Positives = 58/126 (46%), Gaps = 10/126 (7%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+P F ++ + L D L YL V + + G+ + F F NP+F N+
Sbjct 91 IPNFGVTTFVNHPQVSSLLGEEDEEALHYLTKVEVTEFEDIKSGYRIDFYFDENPYFENK 150
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
V SKE + + GD + + T+I W G D+TK R S+ +N +R+ +
Sbjct 151 VFSKEFHLNESGDPS----SKSTKIKWKSGKDVTK--RSSQTQNKASRK----RQHEEPE 200
Query 154 SFFNFF 159
SFF +F
Sbjct 201 SFFTWF 206
> At1g18800
Length=255
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 36/130 (27%), Positives = 52/130 (40%), Gaps = 13/130 (10%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+P FWL+ L+ D+ YL + + + G+S++F F NPFF +
Sbjct 73 IPDFWLTAFLSHPALGELLTEEDQKIFKYLSSLDVEDAKDVKSGYSITFSFNPNPFFEDG 132
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
L+K ++G K T I W EG L V K N R E
Sbjct 133 KLTKTFTFLEEG----TTKITATPIKWKEGKGLANGVNHEKNGNKRA---------LPEE 179
Query 154 SFFNFFENTQ 163
SFF +F + Q
Sbjct 180 SFFTWFSDAQ 189
> At1g74560
Length=256
Score = 57.0 bits (136), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 34/130 (26%), Positives = 54/130 (41%), Gaps = 12/130 (9%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW++ L+ D+ YL + + + G+S++F F +NPFF +
Sbjct 77 IPGFWMTAFLSHPALGDLLTEEDQKIFKYLNSLEVEDAKDVKSGYSITFHFTSNPFFEDA 136
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
L+K ++G K T I W EG L V ++ K E
Sbjct 137 KLTKTFTFLEEG----TTKITATPIKWKEGKGLPNGVNHDDKKG--------NKRALPEE 184
Query 154 SFFNFFENTQ 163
SFF +F + Q
Sbjct 185 SFFTWFTDAQ 194
> 7301638
Length=272
Score = 55.8 bits (133), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 52/193 (26%), Positives = 78/193 (40%), Gaps = 50/193 (25%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKED--GFSVSFVFAANPFF 90
+P FWL VL+ A +S D L L D+R+ RL E F + F F N +F
Sbjct 82 SVPNFWLRVLK--ASYTEFISKRDEKILECLSDIRS--RLYNEPVVKFDIEFHFDPNDYF 137
Query 91 SNRVLSKEI---CMQKQGDD-----AVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTRE 142
+N VL+K C+ D A + K G I W + D TK+
Sbjct 138 TNTVLTKTYFLNCLPDPDDPLAYDGAEIYKCEGCVIDWKQTKDQTKTE------------ 185
Query 143 VRIVKNITSENSFFNFF------ENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIR 196
E SFF FF E+T P+ + AM L ++E+ ++
Sbjct 186 -------NQEPSFFEFFSPPLLPEDTLDPNYCDVNAM-----------LQNDFEVGFYLK 227
Query 197 DIIVPKAIGVYLG 209
+ ++PKA+ + G
Sbjct 228 ERVIPKAVIFFTG 240
> Hs14782699
Length=417
Score = 55.5 bits (132), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 36/128 (28%), Positives = 55/128 (42%), Gaps = 19/128 (14%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW + A L+S ++ L YL + ++ G+ + F F NP+F N+
Sbjct 248 IPGFWGQAFQNHPQLASFLNSQEKEVLSYLNSLEVEELGLARLGYKIKFYFDRNPYFQNK 307
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
VL KE G VV+++ T I W G DL + + + N
Sbjct 308 VLIKEYGCGPSGQ--VVSRS--TPIQWLPGHDLQSLSQGNPENN---------------R 348
Query 154 SFFNFFEN 161
SFF +F N
Sbjct 349 SFFGWFSN 356
> Hs18561397
Length=401
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 33/123 (26%), Positives = 52/123 (42%), Gaps = 10/123 (8%)
Query 37 FWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNRVLS 96
FW+++ + D L YL V + G+ + F F NP+F N +L+
Sbjct 76 FWVTIFVNHPEVSALPGEEDEEALHYLTRVEMTDFDDIKSGYRIDFYFDENPYFENNILA 135
Query 97 KEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSENSFF 156
KE + + GD + + TEI W G DL K ++ + R R+ SFF
Sbjct 136 KEFHLNESGDPS----SKPTEIKWKYGKDLMKRSSQTQNKAHRKRQHE------EPESFF 185
Query 157 NFF 159
+F
Sbjct 186 TWF 188
> Hs22056195
Length=414
Score = 54.7 bits (130), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 42/175 (24%), Positives = 69/175 (39%), Gaps = 38/175 (21%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW++ + +S D L Y+ ++ ++ G F+F NP+F N
Sbjct 250 IPGFWVTAFRNHPQLSPMISGQDEDMLRYMINLEVEELKHPRAGCKFKFIFQGNPYFRNE 309
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
L KE +++ VV ++ T I W+ G D + R RE +
Sbjct 310 GLVKEY--ERRSSGRVV--SLSTPIRWHRGQDPQAHIH-------RNREGNTIP------ 352
Query 154 SFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVYL 208
SFFN+F D + L F R+ A +I+ + P + YL
Sbjct 353 SFFNWF---------------SDHSLLEFDRI------AEIIKGELWPNPLQYYL 386
> Hs11141913
Length=464
Score = 54.3 bits (129), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 42/175 (24%), Positives = 69/175 (39%), Gaps = 38/175 (21%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW++ + +S D L Y+ ++ ++ G F+F NP+F N
Sbjct 300 IPGFWVTAFRNHPQLSPMISGQDEDMLRYMINLEVEELKHPRAGCKFKFIFQGNPYFRNE 359
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
L KE +++ VV ++ T I W+ G D + R RE +
Sbjct 360 GLVKEY--ERRSSGRVV--SLSTPIRWHRGQDPQAHIH-------RNREGNTIP------ 402
Query 154 SFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVYL 208
SFFN+F D + L F R+ A +I+ + P + YL
Sbjct 403 SFFNWF---------------SDHSLLEFDRI------AEIIKGELWPNPLQYYL 436
> 7299983
Length=269
Score = 53.1 bits (126), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 39/177 (22%), Positives = 71/177 (40%), Gaps = 30/177 (16%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+P FW++ + L + CL L + ++ + G+ ++F F NP+F N+
Sbjct 79 IPNFWVTSFINHPQVSGILDEEEEECLHALNKLEVEEFEDIKSGYRINFHFDENPYFENK 138
Query 94 VLSKEICMQKQG--DDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITS 151
VL+KE + ++ + T I W EG +L K + N + R +
Sbjct 139 VLTKEFHLNSAAASENGDWPASTSTPIKWKEGKNLLKLLLTKPYGNKKKRN-------SE 191
Query 152 ENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIGVYL 208
+FF++F + P N E+A +I+D + P + YL
Sbjct 192 YKTFFDWFSDNTDPVND---------------------EIAELIKDDLWPNPLQYYL 227
> HsM4758758
Length=506
Score = 52.8 bits (125), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 45/184 (24%), Positives = 78/184 (42%), Gaps = 14/184 (7%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRAD-KRLRKEDGFSVSFVFAANPFFS 91
G+P +WL VL+ + D L +L DV + + ++ F F NP+F
Sbjct 310 GIPDYWLIVLKNVDKLGPMIQKYDEPILKFLSDVSLKFSKPGQPVSYTFEFHFLPNPYFR 369
Query 92 NRVLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTK------SVRISKQRNLRTREVRI 145
N VL K ++ + D + G EI +G + + +V ++ R T E+ I
Sbjct 370 NEVLVKTYIIKAKPDHNDPFFSWGWEIEDCKGCKIDRRRGKDVTVTTTQSRTTATGEIEI 429
Query 146 VKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPKAIG 205
+ SFFNFF ++P + D L ++E+ ++ D ++ K+I
Sbjct 430 QPRVVPNASFFNFFSPPEIPMIGKLEPRED-------AILDEDFEIGQILHDNVILKSIY 482
Query 206 VYLG 209
Y G
Sbjct 483 YYTG 486
> Hs22063713
Length=258
Score = 52.8 bits (125), Expect = 6e-07, Method: Compositional matrix adjust.
Identities = 44/188 (23%), Positives = 73/188 (38%), Gaps = 32/188 (17%)
Query 2 PLPSAAAPENAAAAEAETAVTPS------------AGYSVATPGLPGFWLSVLEREAHTA 49
PL + A +A+A+ A P A S +PGFW++ +
Sbjct 51 PLEAIQWEAEAVSAQADRAYLPLERRFGRMHRLYLARRSFIIQNIPGFWVTAFLNHPQLS 110
Query 50 RSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNRVLSKEICMQKQGDDAV 109
+S D L YL ++ + G F F +NP+F N+V+ KE + G
Sbjct 111 AMISPRDEDMLCYLMNLEVRELRHSRTGCKFKFRFWSNPYFQNKVIVKEYECRASGR--- 167
Query 110 VAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSENSFFNFFENTQVP-SNR 168
+I T I W+ G + V +N + SFF++F +P ++R
Sbjct 168 -VVSIATRIRWHWGQEPPALVH---------------RNRDTVRSFFSWFSQHSLPEADR 211
Query 169 AIRAMGDD 176
+ + DD
Sbjct 212 VAQIIKDD 219
> Hs20551731
Length=437
Score = 47.0 bits (110), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 31/137 (22%), Positives = 55/137 (40%), Gaps = 19/137 (13%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW++ + + D L Y+ ++ + G F F NP+F N+
Sbjct 275 IPGFWMTAFRNHPQLSAMIRGQDAEMLRYITNLEVKELRHPRTGCKFKFFFRRNPYFRNK 334
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
++ KE ++ G VV ++ T I W G + +R +N
Sbjct 335 LIVKEYEVRSSGR--VV--SLSTPIIWRRGHEPQSFIR---------------RNQDLIC 375
Query 154 SFFNFFENTQVPSNRAI 170
SFF +F + +P + I
Sbjct 376 SFFTWFSDHSLPESDKI 392
> Hs11545835
Length=693
Score = 46.2 bits (108), Expect = 5e-05, Method: Compositional matrix adjust.
Identities = 30/132 (22%), Positives = 55/132 (41%), Gaps = 19/132 (14%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW+ + ++ D YL +++ G+ + F NP+F+N
Sbjct 262 IPGFWVKAFLNHPRISILINRRDEDIFRYLTNLQVQDLRHISMGYKMKLYFQTNPYFTNM 321
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSVRISKQRNLRTREVRIVKNITSEN 153
V+ KE + G +V+ + T I W+ G + + R N + +
Sbjct 322 VIVKEFQRNRSG--RLVSHS--TPIRWHRGQEP---------------QARRHGNQDASH 362
Query 154 SFFNFFENTQVP 165
SFF++F N +P
Sbjct 363 SFFSWFSNHSLP 374
> Hs18561473
Length=225
Score = 45.4 bits (106), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 35/132 (26%), Positives = 61/132 (46%), Gaps = 14/132 (10%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+P F ++ + + +L D L YL V + + G+ + F F N +F N+
Sbjct 91 IPNFGVTTFVKHPQVS-ALLEEDEEALHYLSRVEMTEFEDIKSGYRIDFYFDENLYFENK 149
Query 94 VLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKS------VRISKQRNLRTREVRIVK 147
+LSKE + + GD + + TE+ W G DL K ++ S Q ++ + R +
Sbjct 150 LLSKEFHLNESGDPS----SKSTEMKWKSGKDLMKCSSGKDLMKCSSQ--MQNKASRKRQ 203
Query 148 NITSENSFFNFF 159
+ E SFF +F
Sbjct 204 HEEPE-SFFTWF 214
> Hs17476493
Length=160
Score = 41.6 bits (96), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 25/82 (30%), Positives = 39/82 (47%), Gaps = 4/82 (4%)
Query 47 HTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNRVLSKEICMQKQGD 106
H + L + L YL V + + G+ + F NP+F N+V+SKE + + GD
Sbjct 82 HVSALLGEEEEEALHYLIRVEMTEFENIKSGYRIGFYSDENPYFENKVVSKEFHLNESGD 141
Query 107 DAVVAKTIGTEIHWNEGMDLTK 128
+ + TEI W +LTK
Sbjct 142 PSANS----TEIKWKFRKNLTK 159
> Hs22053134
Length=182
Score = 40.8 bits (94), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 27/104 (25%), Positives = 48/104 (46%), Gaps = 9/104 (8%)
Query 33 GLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRAD-KRLRKEDGFSVSFVFAANPFFS 91
G+P F L+V + + L D L +L+D++ + F + F F N +F+
Sbjct 16 GIPEFRLTVFKNVDLLSDMLQERDEPILKHLKDIKVKFSDACQPMSFVLEFHFEDNEYFT 75
Query 92 NRVLSKEICMQKQGDDA--------VVAKTIGTEIHWNEGMDLT 127
N VL+K M+ + DD+ + G +I W +G ++T
Sbjct 76 NEVLTKTYSMRSEPDDSDPFYFDGPEIMGFTGCQIDWKKGKNVT 119
> Hs13640089
Length=308
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/65 (24%), Positives = 33/65 (50%), Gaps = 0/65 (0%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW +V+ + ++ D L Y+ + ++ + + F +NP+F N+
Sbjct 155 VPGFWANVIANHPQMSALITDEDEDMLSYMVSLEVEEEKHRVHLCKIMLFFRSNPYFQNK 214
Query 94 VLSKE 98
V++KE
Sbjct 215 VITKE 219
> Hs13435135
Length=308
Score = 40.0 bits (92), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 16/65 (24%), Positives = 33/65 (50%), Gaps = 0/65 (0%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW +V+ + ++ D L Y+ + ++ + + F +NP+F N+
Sbjct 155 VPGFWANVIANHPQMSALITDEDEDMLSYMVSLEVEEEKHRVHLCKIMLFFRSNPYFQNK 214
Query 94 VLSKE 98
V++KE
Sbjct 215 VITKE 219
> Hs11999285
Length=308
Score = 38.9 bits (89), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 16/65 (24%), Positives = 32/65 (49%), Gaps = 0/65 (0%)
Query 34 LPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFFSNR 93
+PGFW +V+ + ++ D L Y+ + ++ + F +NP+F N+
Sbjct 155 VPGFWANVIANHPQMSALITDEDEDMLSYMVSLEVEEEKHPVHLCKIMLFFRSNPYFQNK 214
Query 94 VLSKE 98
V++KE
Sbjct 215 VITKE 219
> Hs20558675
Length=123
Score = 35.0 bits (79), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 23/83 (27%), Positives = 39/83 (46%), Gaps = 13/83 (15%)
Query 56 DRACLLYLEDVRADKRLRKED---GFSVSFVFAANPFFSNRVLSKEICMQKQGDDA---- 108
D L +L+D + +L D F + F F N +F+N +LSK M+ + DD+
Sbjct 6 DEPILKHLKDTKV--KLSDADQPMNFVLEFHFEPNEYFTNEMLSKTYRMRSEPDDSDPFS 63
Query 109 ----VVAKTIGTEIHWNEGMDLT 127
+ G +I W +G ++T
Sbjct 64 FDGPEIMGCTGCQIDWKKGKNVT 86
> CE27804
Length=312
Score = 34.3 bits (77), Expect = 0.18, Method: Compositional matrix adjust.
Identities = 21/93 (22%), Positives = 35/93 (37%), Gaps = 5/93 (5%)
Query 31 TPGLPGFWLSVLEREAHTARSLSSGDRACLLYLEDVRADKRLRKEDGFSVSFVFAANPFF 90
T + FW + + ++ L L D+ + GF + F N +F
Sbjct 74 TTKIDNFWQTAFLNHHLLSTAIPEEQEDLLAALRDLEVQEFEDLRSGFKIIMTFDPNEYF 133
Query 91 SNRVLSKEICMQKQGDDAVVAKTIGTEIHWNEG 123
+N V++K +Q + T TEI W E
Sbjct 134 TNEVITKSYHLQSES-----PSTEITEIEWKEN 161
> At3g15450
Length=253
Score = 31.2 bits (69), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 26/80 (32%), Positives = 37/80 (46%), Gaps = 9/80 (11%)
Query 10 ENAAAAEAETAVTPSAGYSVATPGLPGFWLS-VLEREAHTARSLSSGDRACLLYLEDVRA 68
+ A A E +P++ +S TP LPG LS L + A S++ GD A L Y R
Sbjct 6 QKAFAHPPEELNSPASHFSGKTPKLPGETLSDFLSHHQNNAFSMNFGDSAVLAY---ARQ 62
Query 69 DKRLRKE-----DGFSVSFV 83
+ LR+ DG F+
Sbjct 63 ETSLRQRLFCGLDGIYCMFL 82
> At5g46470
Length=1160
Score = 30.8 bits (68), Expect = 2.1, Method: Composition-based stats.
Identities = 13/32 (40%), Positives = 21/32 (65%), Gaps = 0/32 (0%)
Query 178 ARLLFKRLSTEYEMAVVIRDIIVPKAIGVYLG 209
AR LF RLS +++ +V I + + K++ VY G
Sbjct 225 ARALFSRLSCQFQSSVFIDKVFISKSMEVYSG 256
> Hs18576616
Length=203
Score = 30.8 bits (68), Expect = 2.5, Method: Compositional matrix adjust.
Identities = 33/142 (23%), Positives = 66/142 (46%), Gaps = 19/142 (13%)
Query 89 FFSNRVLSKEICMQKQGDDAVVAKTIGTEIHWNEGMDLTKSV----RISKQRNLRTREVR 144
FS ++ KE+C + +AV A T+G I ++ L + + ++ + N
Sbjct 1 MFSYKI-QKELCYEPTAANAVRAGTLGDLISYSILAPLVRLLAGNQKLERHENPSWTIPE 59
Query 145 IVKNITSENSFFNFFENTQVPSNRAIRAMGDDEARLLFKRLSTEYEMAVVIRDIIVPK-- 202
+ IT N +N ++T VPS++ + L ++LS + + V +++ P+
Sbjct 60 PCQKITLAN--YNEIKSTSVPSSKPM---------LSLQQLSGKKQSRVALKERTGPRVW 108
Query 203 -AIGVYLGVAGAESDSEESSDD 223
A+ + L VA A S ++ S+
Sbjct 109 LALFLLLSVASARSTTQHHSEQ 130
> 7301401
Length=677
Score = 30.0 bits (66), Expect = 4.0, Method: Composition-based stats.
Identities = 15/45 (33%), Positives = 21/45 (46%), Gaps = 2/45 (4%)
Query 10 ENAAAAEAETAVTPSAGYSVA--TPGLPGFWLSVLEREAHTARSL 52
E++AA +T P GY + PGFWL EAH + +
Sbjct 612 EDSAAVAKDTVQIPGQGYIIVRFISNNPGFWLYHCHVEAHAVQGM 656
> HsM14719404
Length=854
Score = 29.6 bits (65), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 16/39 (41%), Positives = 18/39 (46%), Gaps = 3/39 (7%)
Query 3 LPSAAAPENAAAAEAETA---VTPSAGYSVATPGLPGFW 38
LP +AP A + T TPS Y TPGLP W
Sbjct 232 LPPPSAPAGRARSSTVTGGEEPTPSVAYVHTTPGLPSGW 270
Lambda K H
0.316 0.130 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 4255059914
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40