bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.24+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: kyva
112,920 sequences; 47,500,486 total letters
Query= Eten_0022_orf1
Length=270
Score E
Sequences producing significant alignments: (Bits) Value
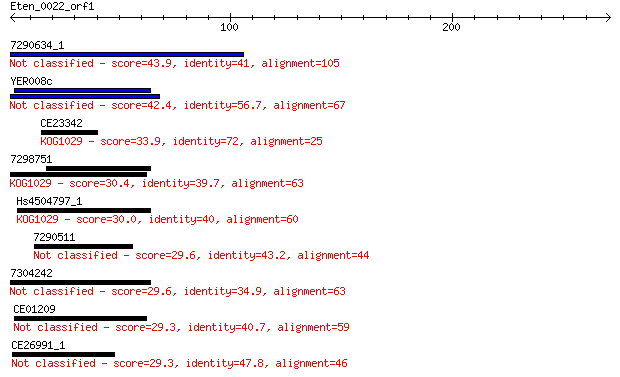
7290634_1 43.9 3e-04
YER008c 42.4 0.001
CE23342 33.9 0.35
7298751 30.4 4.1
Hs4504797_1 30.0 6.0
7290511 29.6 6.6
7304242 29.6 6.8
CE01209 29.3 9.3
CE26991_1 29.3 9.4
> 7290634_1
Length=752
Score = 43.9 bits (102), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 43/105 (40%), Positives = 56/105 (53%), Gaps = 3/105 (2%)
Query 1 EKQKEIESQKEIEKQKEMERQAEMERQRELERQREIEKQEELLKQKELERQKEEEDRLRK 60
E Q E+E+Q E E Q E E Q E+E Q E+E Q E+E Q E+ Q E+E Q E E +
Sbjct 318 EAQPEVEAQPEAEAQPEAEPQLEVEPQPEVESQPEVESQPEVEAQPEVEPQSEVESQPEA 377
Query 61 EREAAAANAAEAEAAPPAEPEPAGAAREEAAAAGPEAEREPAGEA 105
E + AE EA P E P ++ EA + + EREP EA
Sbjct 378 ESHSEPETQAEVEAQPEVESLPEAESQPEAES---QPEREPEVEA 419
> YER008c
Length=1336
Score = 42.4 bits (98), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 36/78 (46%), Positives = 47/78 (60%), Gaps = 21/78 (26%)
Query 3 QKEIESQKEIE----------KQKEMERQAEMERQRELE-----RQREIEKQEELLKQKE 47
QK ++ QKE E KQ+E +RQ E+E QR+LE RQ E+E + KQ E
Sbjct 348 QKRLQLQKENEMKRLEEERRIKQEERKRQMELEHQRQLEEEERKRQMELEAK----KQME 403
Query 48 LERQK--EEEDRLRKERE 63
L+RQ+ EEE RL+KERE
Sbjct 404 LKRQRQFEEEQRLKKERE 421
Score = 39.3 bits (90), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 26/74 (35%), Positives = 51/74 (68%), Gaps = 7/74 (9%)
Query 1 EKQKEIESQKEIE-----KQKEME--RQAEMERQRELERQREIEKQEELLKQKELERQKE 53
++Q E+E Q+++E +Q E+E +Q E++RQR+ E ++ ++K+ ELL+ + +R++E
Sbjct 374 KRQMELEHQRQLEEEERKRQMELEAKKQMELKRQRQFEEEQRLKKERELLEIQRKQREQE 433
Query 54 EEDRLRKEREAAAA 67
+RL+KE + A A
Sbjct 434 TAERLKKEEQEALA 447
> CE23342
Length=1097
Score = 33.9 bits (76), Expect = 0.35, Method: Compositional matrix adjust.
Identities = 18/25 (72%), Positives = 21/25 (84%), Gaps = 0/25 (0%)
Query 15 QKEMERQAEMERQRELERQREIEKQ 39
++E ERQAE+ERQ ELERQR IE Q
Sbjct 339 RQEKERQAEVERQAELERQRIIEAQ 363
> 7298751
Length=1097
Score = 30.4 bits (67), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 19/47 (40%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 17 EMERQAEMERQRELERQREIEKQEELLKQKELERQKEEEDRLRKERE 63
E ER+ + E +R+L+RQREIE ++E +++ELE ++ L K+R+
Sbjct 379 EAERKQQEELERQLQRQREIEMEKEEQRKRELEAKEAARKELEKQRQ 425
Score = 29.3 bits (64), Expect = 8.8, Method: Compositional matrix adjust.
Identities = 19/65 (29%), Positives = 45/65 (69%), Gaps = 4/65 (6%)
Query 1 EKQKEIESQKEIEKQKEMERQAEMERQ----RELERQREIEKQEELLKQKELERQKEEED 56
E +++++ Q+EIE +KE +R+ E+E + +ELE+QR+ E ++ + + ++++E+E
Sbjct 387 ELERQLQRQREIEMEKEEQRKRELEAKEAARKELEKQRQQEWEQARIAEMNAQKEREQER 446
Query 57 RLRKE 61
L+++
Sbjct 447 VLKQK 451
> Hs4504797_1
Length=1221
Score = 30.0 bits (66), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 36/60 (60%), Gaps = 13/60 (21%)
Query 4 KEIESQKEIEKQKEMERQAEMERQRELERQREIEKQEELLKQKELERQKEEEDRLRKERE 63
+EI ++++++KQK ME AE +Q+E ER K ELE+QKEE R +ER+
Sbjct 611 REIHNKQQLQKQKSME--AERLKQKEQER-----------KIIELEKQKEEAQRRAQERD 657
> 7290511
Length=670
Score = 29.6 bits (65), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 19/45 (42%), Positives = 35/45 (77%), Gaps = 2/45 (4%)
Query 12 IEKQKEMERQAEMERQRELERQREIEKQEELL-KQKELERQKEEE 55
+E++K +E++ ER+R L R++++ KQE+LL ++K LER+K +E
Sbjct 387 LEQEKILEQEMVSERERHLTREKQL-KQEKLLEREKHLEREKLQE 430
> 7304242
Length=681
Score = 29.6 bits (65), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 22/63 (34%), Positives = 44/63 (69%), Gaps = 3/63 (4%)
Query 1 EKQKEIESQKEIEKQKEMERQAEMERQRELERQREIEKQEELLKQKELERQKEEEDRLRK 60
E+++E E +E+E +K+ E++AEMERQ+E +R+ E + +L+ + L+ ++E+ RL
Sbjct 458 EREQEKERLQELELRKQEEKEAEMERQKEADRRLE---ETRILEAERLKIEQEDRQRLES 514
Query 61 ERE 63
E++
Sbjct 515 EKQ 517
> CE01209
Length=1039
Score = 29.3 bits (64), Expect = 9.3, Method: Compositional matrix adjust.
Identities = 24/60 (40%), Positives = 36/60 (60%), Gaps = 1/60 (1%)
Query 3 QKEIESQKEIEKQKEMERQAEMER-QRELERQREIEKQEELLKQKELERQKEEEDRLRKE 61
QKE+E ++E +++E ER+ +R Q EL RQ +E +Q E+ RQ E+E R R E
Sbjct 404 QKELEFRREALRKREQEREEHKKRAQEELRRQMTAALEESTRRQAEMNRQIEDEARKRDE 463
> CE26991_1
Length=466
Score = 29.3 bits (64), Expect = 9.4, Method: Compositional matrix adjust.
Identities = 22/48 (45%), Positives = 32/48 (66%), Gaps = 6/48 (12%)
Query 2 KQKEIESQKEIEKQKEMERQAEMERQRELERQREIEKQEEL--LKQKE 47
+ KE+E ++E ++ E +R A MER++E ER R QE+L LKQKE
Sbjct 233 RMKELEKKREKARKDEEKRNAVMERRKEQERVR----QEKLDQLKQKE 276
Lambda K H
0.301 0.116 0.291
Gapped
Lambda K H
0.267 0.0410 0.140
Effective search space used: 5773141498
Database: kyva
Posted date: Jul 3, 2009 9:03 AM
Number of letters in database: 47,500,486
Number of sequences in database: 112,920
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Neighboring words threshold: 11
Window for multiple hits: 40